Recommendations for Fully Vaccinated People
COVID-19 Homepage
CDC’s Role in Tracking Variants
What is CDC doing to track SARS-COV-2 variants?
In the United States, CDC uses genomic surveillance to track emerging SARS-CoV-2 variants that cause COVID-19. CDC established multiple pipelines to connect genomic sequence data from CDC, public health laboratories, and commercial diagnostic laboratories within publicly accessible databases maintained by the National Center for Biotechnology Information (NCBI) and the Global Initiative on Sharing Avian Influenza Data (GISAID).
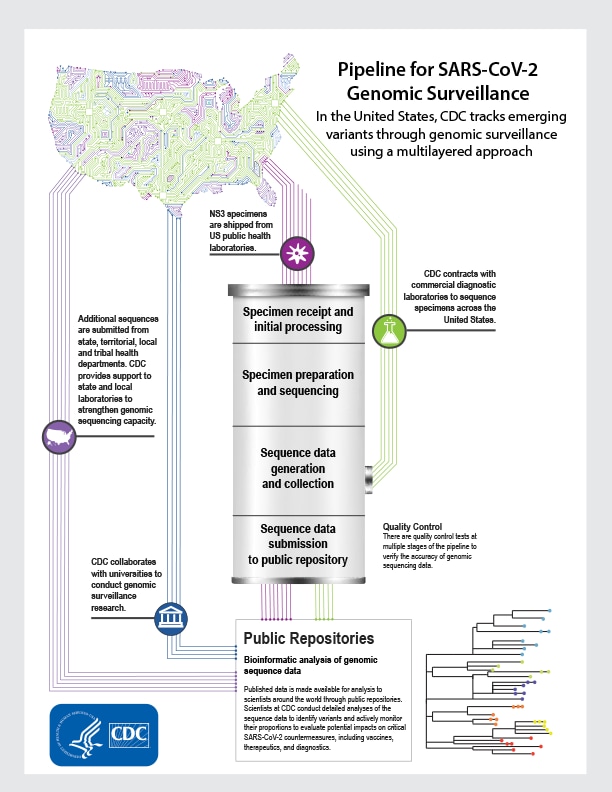
INFOGRAPHIC
Learn how CDC tracks emerging variants through the pipeline for genomic surveillance.
View Infographic [PDF, 3 MB, 1 Page]
As part of the CDC National SARS-CoV-2 Strain Surveillance (NS3) System, public health laboratories ship deidentified specimens to CDC to provide a representative set of viruses for sequencing. There are four main stages in the process of generating SARS-CoV-2 genetic sequence data from these specimens and making it available in public repositories.
Why is genomic surveillance important for public health?
A comprehensive system for SARS-CoV-2 genetic surveillance is important
- Mutations (nucleotide substitutions) occur in viruses and accumulate with continued viral spread; these mutations result in variants that may have different attributes. Genomic surveillance identifies circulating variants to rapidly inform public health response efforts.
- Testing, treatment, and vaccination programs can be improved based on regularly updated surveillance of variants, including updating future vaccines if needed.
- Detection of variants that are more transmissible or cause more severe disease support outbreak preparedness, prevention efforts, and strengthens public health response.
Genomic sequencing process for SARS-CoV-2*
- Specimen receipt and initial processing: Specimens are received and entered into the laboratory information system.
- Specimen preparation and sequencing: SARS-CoV-2 RNA is extracted and converted to complimentary DNA, enriched, and loaded into the next-generation sequencing equipment.
- Sequence data generation and collection: Specimens are sequenced and data is collected from sequencers and initial quality control steps are taken. The raw data is processed and turned into sequence data. At this point, a parallel process begins where sequence data collected from commercial laboratories are integrated with CDC databases for processing.
- Sequence data submission to public repositories: Scientists conduct quality control steps. Sequences not initially accepted by public repositories are analyzed and potentially re-sequenced for resubmission. Published data are made available to scientists around the world through public repositories.
Bioinformatic analysis of Genomic Sequence Data
Published data are available to scientists around the world through public repositories. CDC regularly collects genomic sequence data from multiple sources to support national surveillance. Commercial laboratories transfer genomic sequence data to CDC at step 3 where it is processed and submitted for publishing. Public health laboratories, research laboratories, and universities submit data directly to public repositories. Scientists at CDC conduct detailed analyses of the sequence data to identify variants and actively monitor their proportions to evaluate potential impacts on critical SARS-CoV-2 countermeasures, including vaccines, therapeutics, and diagnostics.
CDC encourages state public health laboratories to “tag” the sequences they generate and publish in public databases to include their output in CDC analyses. The use of standard and consistent tagging information on the submitted sequences improves our ability to search, analyze, and share the data generated across the Unites States.
* There are quality control tests at multiple stages of the pipeline to verify the accuracy of genomic sequencing data.
Leading the National SARS-CoV-2 Strain Surveillance (NS3) system
The NS3 program collects, analyzes, and shares information about the genetic diversity of SARS-CoV-2, the virus that causes COVID-19. The program provides a comprehensive and population-based surveillance system for the United States to track virus evolution over time and identify emerging variants that may affect the performance of diagnostics, therapeutics, or vaccines, or that impact the transmissibility of SARS-CoV-2 or severity of COVID-19. A notable strength of NS3 is the regular collection of specimens from across the United States to support variant characterization efforts, which provides important data to inform public health decision-making.
Through partnerships with state and local public health agencies CDC requests up to 750 total specimens per week from all states and jurisdictions for sequencing and further characterization of representative viruses. The NS3 program has three main goals:
- National virus monitoring: US public health laboratories send representative SARS-CoV-2–positive clinical specimens to CDC on a weekly or bi-weekly basis to support federal efforts to sequence, genetically analyze and phenotypically characterize the viruses circulating among our population over time. This also supports a repository of public SARS-CoV-2 sequence data and specimens.
- Enhanced Surveillance: Since the emergence of SARS-CoV-2, national and global sequencing efforts have identified changes in the SARS-CoV-2 genome as a result from transmission and evolution in humans and animals. These genetic changes can affect many aspects of public health including transmission, disease severity, diagnostics, therapeutics, and vaccines. As part of NS3 enhanced surveillance, CDC may request additional specimens from public health laboratories to investigate variants of interest, variants of concern, or other specified viral classifications, or cases of vaccine breakthrough.
- Virus characterization: Based on genomic analyses, SARS-CoV-2 variants are isolated from positive specimens provided by US public health laboratories. These isolated viruses are evaluated in CDC laboratories to understand their potential impact on current vaccines, treatments, and diagnostics, and their overall risk to public health.
As laboratories have scaled up sequencing capacity, CDC works to improve and build additional technical infrastructure and workflows to ensure efficient sequence data submission to public repositories, which store publicly accessible sequence data for scientists to include in analyses. Next-generation sequencing is a multi-step process that involves both laboratory and bioinformatic workflows. The time from specimen receipt at CDC to having an assembled sequence ready for submission to public databases is approximately 10 days. A similar timeline often applies to state, local, academic, and commercial partners.
The genetic sequence data generated by CDC and state and local public health laboratories are submitted to publicly accessible databases maintained by the National Center for Biotechnology Information (NCBI) and the Global Initiative on Sharing Avian Influenza Data (GISAID). Integration of surveillance across the United States maximizes the sequencing capacity, expertise, and data that is available to inform public health decision making.
Partnering with commercial diagnostic laboratories
In addition to the NS3 program, CDC contracted with large commercial diagnostic laboratories to sequence specimens from across the United States. These contracts provide consistent access to SARS-CoV-2 sequence data across the country to supplement existing public health sequencing efforts.
Collaborating with universities
CDC has funded 29 universities to conduct genomic surveillance research in collaboration with public health agencies. The studies provide deeper insights into viral genomics and molecular epidemiology within the various regions across the country. These insights include:
- Genomic surveillance and outbreak investigation
- Vaccine breakthrough
- Patient outcomes and risk factors for severe disease
- Transmission on or around schools and institutes of higher education
These collaborations further develop and build upon bioinformatic and sequencing capabilities around the country, which are necessary components of a comprehensive public health infrastructure.
Supporting state, territorial, local, and tribal health departments
Since 2014, CDC’s Advanced Molecular Detection program has integrated next-generation sequencing and bioinformatics capabilities into the US public health system. Many state and local health departments use genomic sequencing as part of their response to the COVID-19 pandemic. Sequencing conducted by public health departments can help provide a better understanding of local epidemiology and transmission, and these data can help investigate clusters of disease in various settings, such as healthcare facilities. State and federal public health departments support local investigations, conduct studies, and make genomic data available to public databases. To further support these efforts, over $250 million has been released to public health departments since the start of the COVID-19 pandemic.
Leading the SARS-CoV-2 Sequencing for Public Health Emergency Response, Epidemiology, and Surveillance (SPHERES) consortium
Since early in the COVID-19 pandemic, CDC has led a national consortium of laboratories sequencing SARS-CoV-2, known as SPHERES. The SPHERES consortium consists of more than 250 institutions and includes almost 1,200 scientists from across the United States, including academic centers, industry, non-governmental organizations, and public health agencies.
These efforts help:
- Coordinate sequencing and data analysis across the country
- Encourage collaboration and the use of consistent data standards, protocols, and best practices
- Ensure that generated genomic data are of the highest quality and provide the most impact for public health and the broader research community
Virus Characterization
CDC analyzes the available genomic sequence data and provides weekly updates:
- Published SARS-CoV-2 sequences
- SARS-CoV-2 variant proportions
- Proportions of SARS-CoV-2 variants with substitutions of therapeutic concern.
These analyses drive the selection and prioritization of a subset of representative viruses for further characterization. Virus characterization efforts may include evaluation of the ability of antibodies generated after a previous infection or vaccination to neutralize circulating viruses, the susceptibility of viruses to authorized treatments, and laboratory studies to assess virus transmissibility and pathogenesis. Virus isolates are shared with public health, academic, federal, and commercial partners through BEI Resources.
To receive email updates about COVID-19, enter your email address: