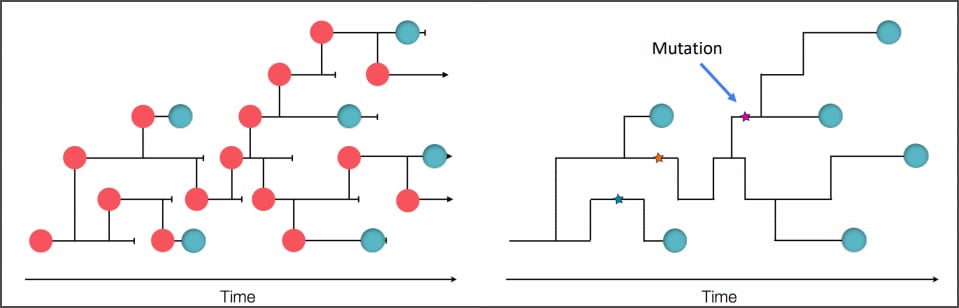
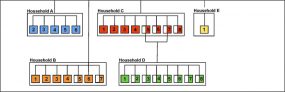
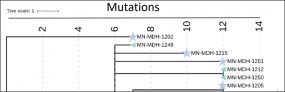
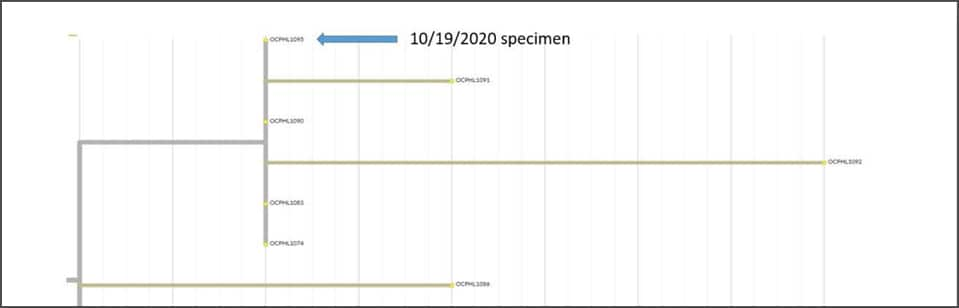
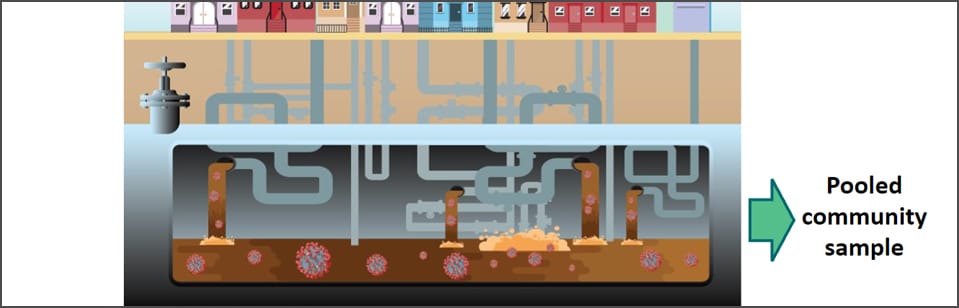
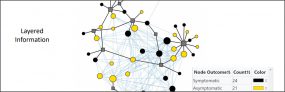
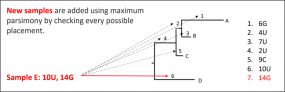
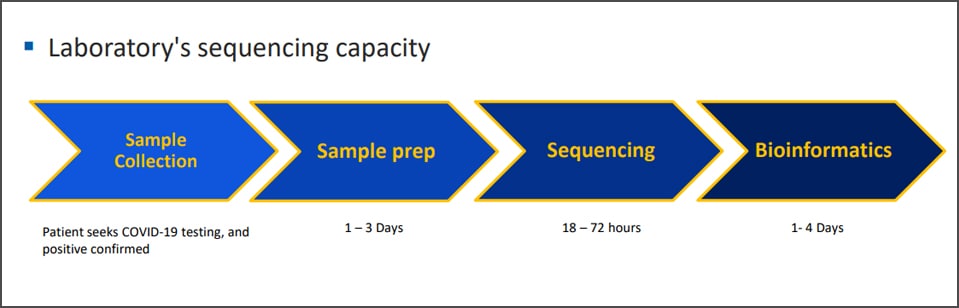
COVID-19 Genomic Epidemiology Toolkit
The Office of Advanced Molecular Detection presents this toolkit to address topics related to the application of genomics to epidemiologic investigations and public health response to SARS-CoV-2. The COVID-19 Genomic Epidemiology Toolkit is meant to further the use of genomics in responding to COVID-19 at the state and local level.
Each module includes a dedicated survey to inform future training development. We value your input.
portrait icon Meet the developers
Overview: CDC’s Dr. Greg Armstrong gives an introduction to the COVID-19 Genomic Epidemiology Toolkit and describes the role for genome sequencing in public health.
Posted: 01/08/21
Presenter: Gregory L. Armstrong, MD
Director, Advanced Molecular Detection Program, CDC
More modules and materials will be added to this toolkit, so please check back for updates or subscribe to the mailing list.