Detecting Outbreaks with Whole Genome Sequencing
For over 20 years, public health laboratories have used a DNA fingerprinting technology called Pulsed-Field Gel Electrophoresis (PFGE). In recent years, a set of new technologies have revolutionized our ability to decode DNA. Whole genome sequencing (WGS) gives us a much more detailed DNA fingerprint. In public health, this is transforming how epidemiologists and laboratory scientists approach the detection and investigation of outbreaks. This allows public health agencies across the US to detect outbreaks sooner, including many outbreaks that would previously gone undetected.
Salmonella is one of the most common causes of foodborne illness. Salmonella serotype Enteritidis are responsible for about one in six Salmonella infections in the United States. To show how this technology is used, we’ve taken 149 cases of Salmonella serotype Enteritidis from the seven states in this region in 2018 and represented each case by a gray dot.

During the one year represented above, several outbreaks were identified, either in real time or retrospectively. The cases in those outbreaks are shown in color. Finding the outbreaks is much easier and faster if they can be sorted from the non-outbreak cases—the cases shown in grey.
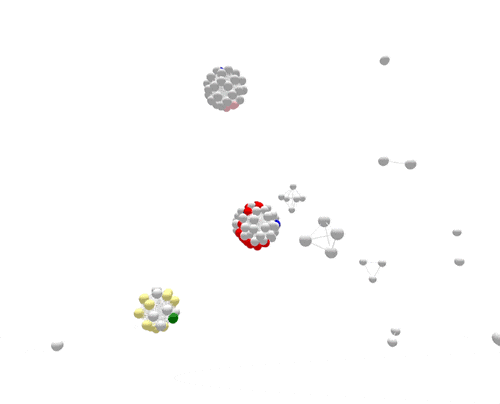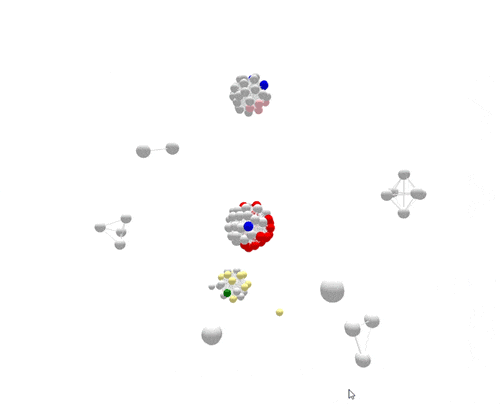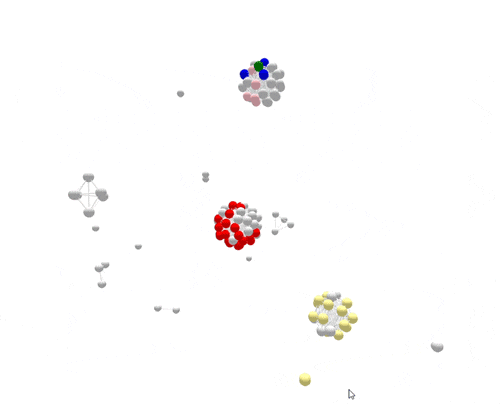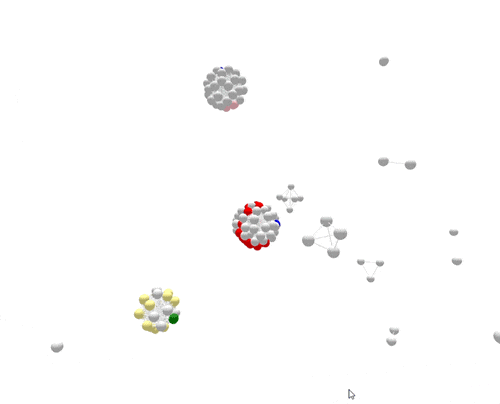
For the most part, PFGE correctly sorts cases that are related and part of the same outbreak from other unrelated cases. However, if you focus on the largest outbreak here, represented in red, you’ll notice that many grey non-outbreak cases are mixed in with the outbreak cases, complicating the investigation.
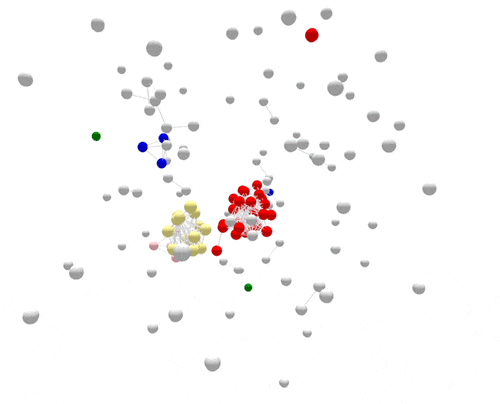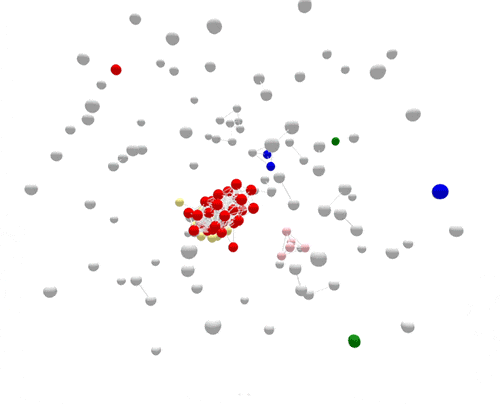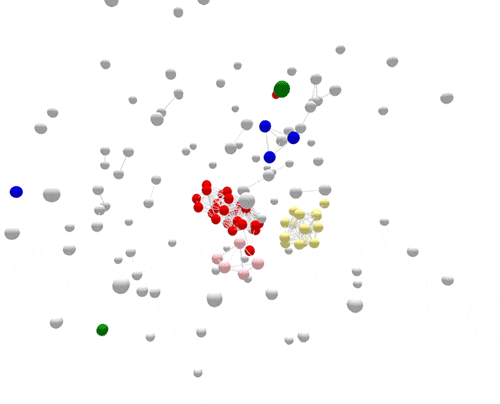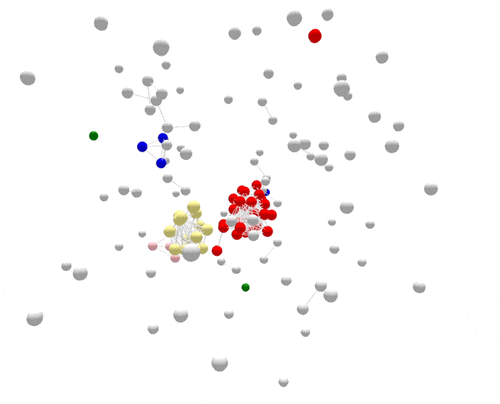
Using WGS, the outbreak cases are much more tightly clustered, and stand out clearly from the disconnected, non-outbreak cases. The largest (red) cluster here was originally detected among a small number of patrons at two different restaurants in two separate states. But WGS showed that both clusters of disease were actually part of a larger outbreak, involving several patients who had not been to either restaurant.
After epidemiologic data suggested the cause of the outbreak—shell eggs—whole-genome sequencing was used to verify the source. Salmonella was found in the implicated eggs, and the DNA fingerprint of the egg isolate matched the outbreak, confirming the attribution and leading to a nationwide recall.
In the US, through the federally funded AMD Program, public health agencies are applying next-generation sequencing in almost every area of infectious disease public health.
In food safety, CDC is working with FDA, USDA, NIH and state and local public health agencies to intervene more quickly in outbreaks and to better understand how to prevent pathogens from getting into the food system in the first place.
In flu, sequencing is enabling faster, more effective characterization of viruses to better understand how they emerge and to improve vaccines
In viral hepatitis, sequencing has proven invaluable in investigating outbreaks.
These are but a few of the areas where the application of sequencing is improving public health surveillance and outbreak response.