Building AMD Capacity in State and Local Laboratories
A critical focus of the AMD program is accelerating the implementation of AMD technology in all state and large local health departments. CDC’s AMD program works with other experts at CDC to ensure the U.S. has the technology and infrastructure to protect Americans from infectious diseases, including emerging threats that may not have been possible to track using older technologies. The CDC AMD program develops and pilots next-generation diagnostics and protocols with and for CDC and state and local public health labs. These tools are then leveraged by programs across CDC and brought to scale in public health labs nationwide.
The following examples show the potential impact of AMD technologies when leveraged through CDC’s programs:
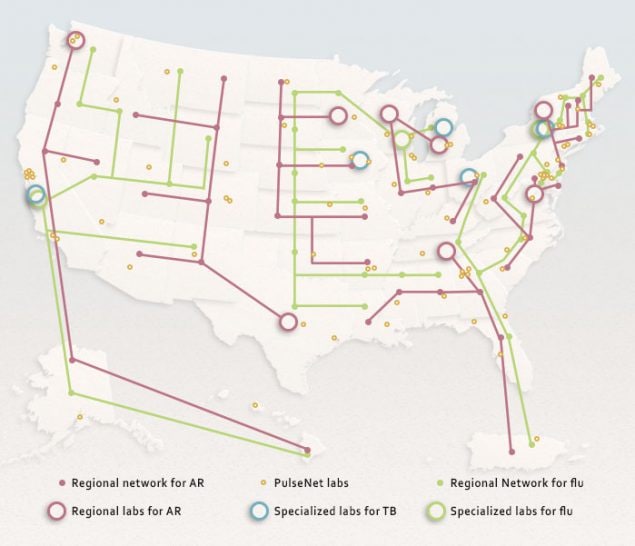
Through AMD funding, networks of regional and state laboratories are applying next-generation sequencing to protect Americans from many infectious disease threats including Antibiotic Resistance (AR), foodborne disease (PulseNet), tuberculosis (TB), and influenza (flu).

In 2015, AMD-funded DNA sequencing tools were used to confirm cases belonging to a Listeria outbreak across four states, including Oklahoma. Ten people were infected and hospitalized and there were three deaths reported. The outbreak was linked to ice cream from Blue Bell Creameries. Whole genome sequencing helped detect this Listeria outbreak in a food source that would likely never have been identified without AMD methods.
AMD-funded DNA sequencing tools helped public health laboratory and disease detectives solve a 2016 E. coli outbreak linked to flour. The outbreak impacted people in 24 states, including Washington, and resulted in 63 illnesses and 17 hospitalizations. As a result of this outbreak investigation, public health agencies updated their warnings against eating raw dough or batter to include the possibility of getting sick with E. coli from uncooked flour. While this was not the first outbreak suspected to be related to E. coli contamination in flour, DNA sequencing in concert with epidemiologic investigation, allowed us to make the link for the first time.

CDC laboratory scientists used AMD technologies to combat one of the largest outbreaks of HIV in recent years. During the outbreak, more than 200 people were infected with HIV and over 90% were co-infected with hepatitis C. By combining AMD technology with traditional on-the-ground epidemiology and laboratory tools, CDC determined that the outbreak was limited to one county, enabling public health officials to focus limited resources among people at greatest risk and most in need of testing and treatment services.

California implemented DNA sequencing in its state lab and a few of its county laboratories. The first area where state labs integrate AMD technologies is often foodborne disease surveillance. AMD helped to support the infrastructure at CDC and in state and local health departments to move from a 20-year-old technology (PFGE) to state-of-the-art whole-genome sequencing. Use of this technology for Listeria, a common cause of foodborne illness, shows that it detects clusters of illness sooner, links more outbreaks to specific foods, and detects clusters of illnesses that would not have been detected by the older methods. As of January 2018, all states switched to DNA sequencing to detect and track Listeria. This system is rapidly expanding to include all foodborne bacteria.

The Minnesota Department of Health partners with CDC and the Mayo Clinic to better identify tick-borne Borrelia infections across the United States using AMD methods. Some sub-groupings of the Borrelia bacterial species cause human disease. One grouping causes relapsing fevers and another causes Lyme disease, the most common tick-borne disease in the United States. In 2017, investigators discovered a bacterium, Borrelia johnsonii, in a human for the first time. Previously, this bacterium was thought to be found only in bat ticks. A better understanding of the type and geographic distribution of Borrelia species infecting humans will improve clinical recognition of disease, laboratory diagnosis, and prevention.
Through AMD investments, CDC hepatitis experts developed a custom NGS approach for sequencing hepatitis C virus (HCV) and launched the Global Hepatitis Outbreak and Surveillance Technology (GHOST). GHOST is a cloud-based system that integrates NGS data with bioinformatics and information technologies. Regardless of computer expertise, GHOST users can conduct cost-effective, accurate HCV molecular surveillance. CDC is currently training states on how to use AMD methods and upload NGS sequence data into GHOST. Because of this training, participating state public health laboratories used GHOST in 2017 to identify outbreaks on their home turf.
In April 2017, Tennessee Department of Health (TDH) identified a cluster of seven recent HIV cases. Because the laboratory and epidemiology staff had trained on and pilot tested CDC’s GHOST technologies in 2016, they used this powerful AMD tool to investigate the cluster. Among the 22 cases (7 from the HIV cluster and 15 from outreach testing), GHOST found 4 linkages – all from outreach testing. In this cluster, two people were linked to patients who had been tested during the massive testing that took place during the state’s 2016 GHOST pilot project.
In 2017, Michigan Department of Health and Human Services (MDHHS) epidemiologists suspected they were seeing a cluster of HCV infections, but traditional surveillance and investigation did not reveal connections between the cases. MDHHS laboratory and epidemiology staff had recently attended training on CDC’s GHOST system. They turned to AMD methods to sequence the DNA of the virus from those infected and used GHOST to corroborate genetic relatedness of HCV among those infected, verifying a cluster of HCV infections.
The New York State and Westchester County Departments of Health investigated an alternative medicine practice following diagnosis of HCV infection in some of its patients. The state’s Wadsworth Center laboratory performed next-generation sequencing and GHOST analysis on specimens from five HCV-positive patients who had received intravenous infusions at the practice. GHOST analysis identified a single transmission cluster linking all five patients, providing critical evidence that patients were infected with HCV through procedures conducted at the medical practice.

The AMD Program collaborates with the Antibiotic Resistance (AR) Solutions Initiative to scale up the use AMD technologies by public health laboratories nationwide. AR investigations highlight the importance of using AMD technologies to detect emerging resistance, identify global resistance threats, and prevent the spread of resistance in the United States.
Hawaii
The Hawaii Department of Health (HDOH) is a critical sentinel site for monitoring antibiotic-resistant gonorrhea using AMD next-generation sequencing. In collaboration with CDC, the HDOH rapidly identified strains of gonorrhea from seven patients that had elevated resistance to CDC’s recommended treatment regimens. This allowed disease investigation specialists to ensure the patients received appropriate treatment and that there was follow-up with individuals who had been in contact with them—containing any spread of this potentially resistant strain of gonorrhea.
Michigan
The Michigan public health laboratory is working with CDC’s Tuberculosis and Antimicrobial Resistance programs to sequence all newly diagnosed TB cases (estimated to be 4,000-9,000 based on past years) for antimicrobial resistance. AMD technologies are shedding valuable light on how antimicrobial resistance is emerging and spreading—revealing the genes that cause resistance to antibiotics and often showing their geographic origin. Understanding the emergence and spread of antibiotic-resistant pathogens is critical for starting to contain them.