AMD Activities: 2016

In 2016, an outbreak of E. coli infections made more than 60 people in 24 states sick and almost a third were hospitalized. Sick people reported eating raw dough or batter and several children played with raw dough at restaurants. The U.S. Food and Drug Administration identified E. coli in bags of flour, which were produced at a single U.S. facility. Investigators used whole genome sequencing, an AMD method, to include ill people in the outbreak who would have been excluded using traditional methods and to confirm that the E. coli making people sick was closely related to the E. coli in the flour. The flour producer recalled more than 45 million pounds of flour.
Brugia malayi is one parasitic organism responsible for the disease lymphatic filariasis.
Diseases caused by parasites are notoriously difficult to identify. Even when scientists can acquire sequence data, parasitic genomes are so large that they require specialized computer programs to decipher. But AMD is helping CDC scientists explore the complex parasitic genome in new and faster ways. A new detection method developed by CDC scientists has the potential to revolutionize parasitic testing by selectively detecting all parasite DNA without interference from human DNA in blood, feces, tissue, or fluids. This will make it easier to identify not just which parasite made someone sick, but also important genetic information about it. All of this in one test. Once the test is validated, CDC scientists will make it available to other researchers, who could apply it to additional pathogens and disease processes.
The dwarf tapeworm, Hymenolepis nana.
In 2013, doctors in Colombia asked CDC to help diagnose unusual tumors from a man infected with HIV. While the tumors resembled cancer, their cells were much smaller than human cancer cells. After performing dozens of tests, CDC identified the cancer-like cells as being from the dwarf tapeworm, Hymenolepis nana, and AMD proved the mutations were in the tapeworm genome. This is the first time scientists have seen a tapeworm’s cancer cells take root in a person. Researchers suspect the cancer transferred to the patient because his immune system was weakened by HIV infection. Because HIV is a global threat and the dwarf tapeworm is found worldwide, similar cases may occur. Thanks to work through AMD, clinicians will be aware and able to spot additional cases of this rare disease.

Recently a new pathogen that causes Lyme disease, Borrelia mayonii, was found in blacklegged ticks in Minnesota and Wisconsin.
In 2016, Mayo Clinic and CDC scientists described a novel cause of Lyme borreliosis in North America, the bacterium Borrelia mayonii. Using AMD, CDC scientists sequenced the full genome of the newly discovered bacterium, providing a critical foundation for diagnostic development. AMD methods are also being used to broadly detect bacteria that may be causing illness in patients with suspected tickborne disease. This approach has already led CDC researchers to discover additional novel tickborne bacterial pathogens, not associated previously with human infection.
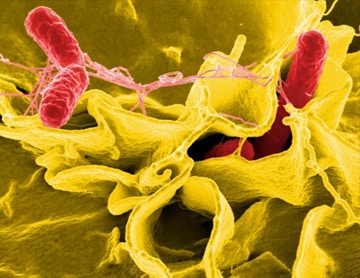
CDC is using AMD methods to develop new tests that can identify foodborne bacteria, such as Salmonella, directly from patient specimens without the need to culture the bacteria first.
Culture-independent diagnostic tests (CIDTs) are the future of foodborne disease detection in hospitals and doctors’ offices. These tests rapidly identify pathogens without the need to culture (grow) it first, yielding a faster diagnosis. But CIDTs pose a problem for public health investigators. The PulseNet surveillance system relies on isolates to identify outbreaks. Public health laboratories need isolates to determine the bacteria’s DNA fingerprint and upload in PulseNet databases. Fortunately, CDC is developing tools to identify and extract disease-causing bacterial DNA from stool samples. These tools will continue to identify and isolate circulating strains of foodborne pathogens.
