CDC launches Traveler-based SARS-CoV-2 Genomic Surveillance Program
Early warning detection system for new SARS-CoV-2 variants active at four US International airports
US airports serve more than 1 billion travelers each year. Public health professionals at these locations are on the front lines to detect new lineages of SARS-CoV-2, the virus that causes COVID-19, that enter the United States through arriving travelers. Travelers are an especially important group to consider when tracking new and emerging infectious diseases. That’s because they are mobile, have potential to be exposed during travel, and can spread diseases from one place to another in a short amount of time.
To detect imported emerging infectious diseases, health officials have traditionally looked at travelers who report to health clinics after symptom onset. It is uniquely challenging to detect SARS-CoV-2 because people infected with the virus can often spread it before they have symptoms. Sometimes people get infected and never develop symptoms at all, but they can also spread the virus to others.
By the time variants are detected in collected samples from people seeking testing, there is often widespread community transmission.
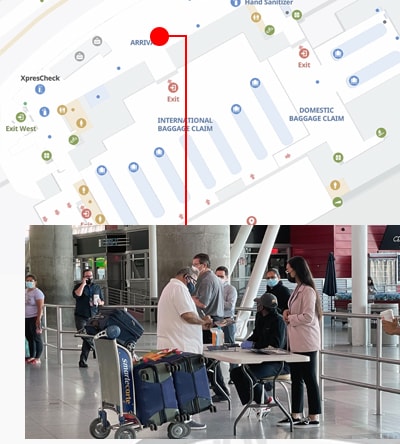
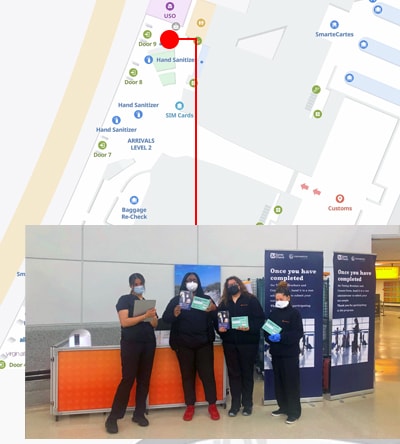
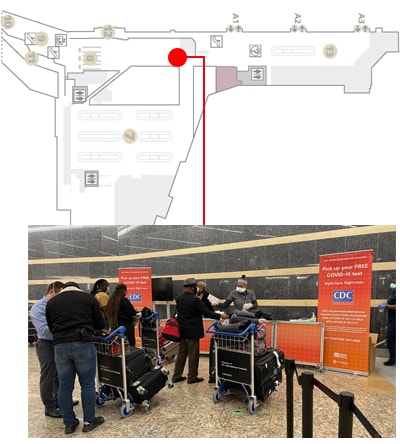
An innovative new program, CDC’s Traveler-based SARS-CoV-2 Genomic Surveillance Program, leverages multiple strategies, including pooled sample collection from arriving passengers, to detect new and rare SARS-CoV-2 lineages early.
The public-private initiative, led by CDC’s Travelers’ Health Branch in partnership with XpresCheck (an airport-based company that offers COVID-19 testing) and Concentric by Ginkgo (a network of over 60 laboratories with genetic sequencing capability), was supported by CDC’s Advanced Molecular Detection (AMD) Program when the pilot program launched in September 2021.
“One role of the AMD Program is supporting innovation and the adoption of novel technologies,” says Gregory Armstrong, MD, Director of the AMD Program. “So, when the Division of Global Migration and Quarantine (DGMQ) approached us last year with this idea, we were intrigued, and agreed to fund the pilot.”
The idea for the program evolved in March 2021. As the COVID-19 pandemic continued to escalate and the Delta variant was emerging in India, Cindy R. Friedman, MD, Chief of the Travelers’ Health Branch in DGMQ, considered how to detect any new SARS-CoV-2 lineages global air travelers might be transporting into the United States.
At around the same time, two companies, XpresCheck and Concentric by Ginkgo were hoping to make a public health contribution to address the global pandemic. So, Friedman reached out and suggested a collaboration. The three entities would create a voluntary SARS-CoV-2 genomic surveillance program, led and funded by CDC, for international travelers arriving in the United States.
The partners launched an eight-week pilot program in September 2021 that included seven flights from India, arriving at three US airports: John F. Kennedy International in New York, Newark Liberty International in New Jersey, and San Francisco International in California.
“We wanted to achieve several things: to get the platform set up at three airports, gauge the level of participation, and determine if we could detect variants using pooled sampling,” says Friedman. “We achieved our goals and showed proof of concept.”
In late November 2021, as the pilot was reaching week eight, news emerged of the Omicron variant spreading in Southern Africa. In response, the program rapidly scaled-up to add Hartsfield-Jackson Atlanta International Airport and to include travelers on flights from Africa, Europe, and South America. During the first weeks after the expansion, the program was the first to report the first BA.2 variant in the United States (seven days before it was reported elsewhere) and the first BA.3 variant (43 days earlier) in North America. With the expansion of the program, samples have been collected from travelers arriving from more than 15 countries on five continents.
Select positive samples are sent to CDC for viral characterization. This critical step provides vital information about new variants that can help health officials adjust their public health prevention strategies. The program has been a successful addition to CDC’s public health tool kit.
Genomics is a key tool to advance public health for CDC. In 2021, Congress included 1.75 billion dollars for genomics through the American Rescue Plan with two very broad priorities:
- To accelerate sequencing of SARS-CoV-2 and
- To expand the use of genomics within the U.S. public health system.
“Pathogen genomic sequencing is widely applicable in public health, as the current pandemic demonstrates,” says Armstrong, stressing that the earlier a new SAR-COV-2 variant is detected and can be characterized, the quicker scientists can take appropriate public health action.
“This Traveler-based SARS-CoV-2 genomic surveillance program is a model of pathogen detection that can be used as an early warning system for pandemics and other outbreaks affecting international travelers,” says Friedman.