
Stool Specimens – Molecular Diagnosis
Microscopic examination is still considered the “gold standard” for the diagnosis of parasitic diseases. If an unequivocal identification of the parasite can not be made, the stool specimen can be analyzed using molecular techniques such as polymerase chain reaction (PCR). PCR amplified fragments can be analyzed by using restriction fragment length polymorphisms (RFLP) or DNA sequencing if further characterization is needed.
Specimen Collection
If PCR is being requested on a stool specimen, the specimen must be collected in a preservative that is compatible with molecular detection. Fixatives/preservatives with acceptable performance for PCR include TotalFix, Unifix, modified PVA (Zn- or Cu-based), and Ecofix. Stool specimens in these preservatives can be stored and shipped at room temperature. Alternatively, stool specimens can be collected in a clean vial and kept unpreserved; however, these specimens must be stored cold or frozen and shipped either refrigerated (4°C) or frozen (shipped with dry ice). Trichrome stained smears (for E. histolytica/E. dispar) or acid-fast smears (for C. parvum or C. cayetanensis) should accompany the stool specimen when requesting PCR for any of these protozoa. All stained smears will be read first and if an identification of the parasite can be made, PCR will not be performed. Click here for more information about shipping stool specimens to CDC.
Fixatives/preservatives that are not recommended for molecular detection include formalin, SAF, LV-PVA, and Protofix.
For specific applications or when commercial fixatives are not an option, the stool can be mixed in potassium dichromate 2.5% (1:1 dilution) or in absolute ethanol (1:1 dilution) and shipped refrigerated.
DNA Extraction
It is necessary to extract DNA from the stool specimens for PCR detection. Click to view the DNA extraction protocols recommended for molecular diagnosis of intestinal parasites.
PCR Analysis
Molecular detection of Cyclospora cayetanensis, Entamoeba histolytica, and E. dispar is performed at CDC by both conventional PCR and real-time PCR. Conventional PCR is available for microsporidia. Click on the links above to learn more about the specific tests and to view analysis of PCR results for the respective parasites listed.
Conventional PCR:
DNA preparations extracted from fecal samples are tested by PCR with diagnostic primers. Amplified DNA fragments are electrophoretically resolved on an agarose gel for analysis of results.
Real-Time PCR
In real-time PCR, the DNA amplification is monitored by measuring the fluorescence signal generated in the reaction vessel. The fluorescence signal is measured every cycle and is proportional to the amount of accumulated PCR product.
The real-time PCR assays for parasite detection at CDC use either the DNA-binding dye SYBR Green or sequence-specific TaqMan probes as fluorescence detection mechanisms. Probe-based assays have the advantages of high specificity and the possibility to detect multiple targets in the same vessel by combining probes labeled with dyes with different fluorescent spectra. Assays using SYBR Green can be easier and less expensive, but caution should be exercised since all double-stranded DNA is detected, including primer-dimers and other PCR artifacts. The specificity of these assays can be improved by including a dissociation (melting) curve analysis. This helps to distinguish the correct product from possible artifacts, thus avoiding false-positive results. See illustrations for details about the fluorescence detection mechanisms using SYBR Green and TaqMan probes.
For additional information on molecular diagnosis using stool specimens, call the Division of Parasitic Diseases at (404) 718-4120.
Real-time PCR Illustrations
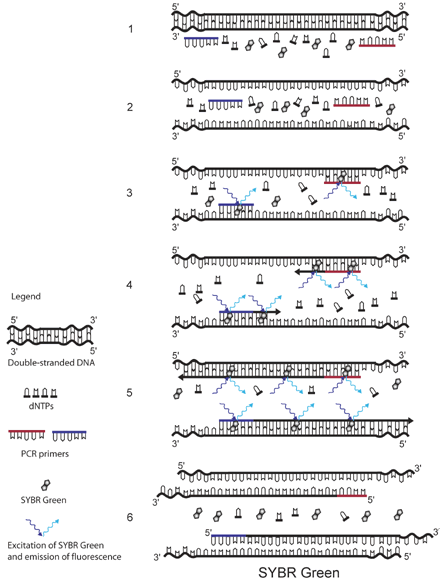
The principle of SYBR Green detection in real-time PCR is outlined in the figure above.1 The fluorescent dye SYBR Green is added to the PCR mixture (1). SYBR Green is a DNA binding dye that fluoresces strongly when bound to double-stranded DNA. At the start of the reaction, very little double stranded DNA is present, and so the fluorescent signal detected by the thermocycler is low (3). As the reaction proceeds and PCR product accumulates, the amount of double-stranded DNA increases and with it the fluorescence signal (4-5). The signal is only detectable during annealing and extension, since the denaturation step contains predominantly single-stranded DNA (6).
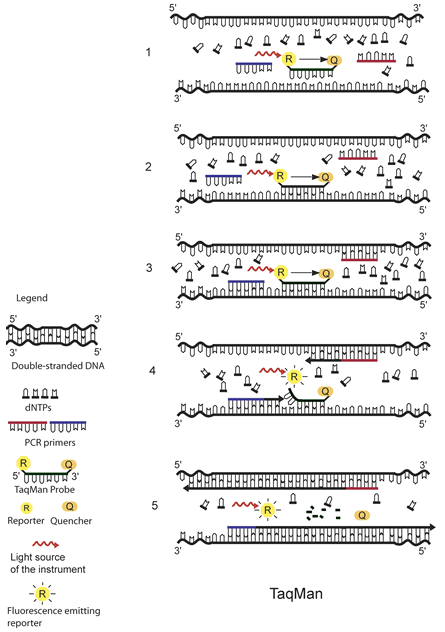
The principle of TaqMan real-time PCR is depicted in the schematic above.1 The TaqMan probe is designed to be complementary to a specific sequence spanned by the PCR primers. The TaqMan probe has a reporter dye at its 5́ end and a quencher dye at its 3́ end. As long as the probe is intact and the reporter and the quencher dyes are in close proximity, no fluorescence signal is emitted due to the quenching effect (black arrow in 1, 2, and 3) (1). After the annealing of the TaqMan probe (2) and the primers (3), the primers are extended by the DNA polymerase. As the polymerase reaches the TaqMan probe, it uses its exonuclease activity to remove the probe one nucleotide at the time (4). This releases the reporter from the proximity of the quencher and allows for the release of a fluorescence signal from the reporter (5).
References
- da Silva A, Pieniazek N. Latest Advances and Trends in PCR-based Diagnostic Methods. In: Dionisio D, editor. Textbook-Atlas of Intestinal Infections in AIDS. Springer; 2003. p. 397-412.
DPDx is an educational resource designed for health professionals and laboratory scientists. For an overview including prevention, control, and treatment visit www.cdc.gov/parasites/.