Antifungal Susceptibility Testing Yeasts Using Gradient Diffusion Strips
About
Gradient diffusion strips (sometimes referred to by the trade name Etest™) are an accurate, inexpensive methodology for testing yeasts for antifungal susceptibility.
1.0 Purpose/Principle
This document describes antifungal susceptibility testing of yeasts using gradient diffusion strips. Currently, there are at least three commercial manufacturers of gradient diffusion strips that measure antifungal susceptibility: Biomerieux, Liofilchem and Himed. This method of antimicrobial testing is based upon a continuous concentration gradient of drug infused on a plastic non-porous strip. The antifungal agent diffuses into an agar plate and allows an accurate determination of MIC values based on elliptical growth around the antifungal gradient.
3.0 Related Documents
| Title | Document |
|---|---|
| Reference Method for Broth Dilution Antifungal Susceptibility Testing for Yeasts | Clinical and Laboratory Standards Institute standard M27, 4th Ed. |
| Performance Standards for Antifungal Susceptibility Testing of Yeasts | Clinical and Laboratory Standards Institute document M60, 2nd Ed. |
| Gradient Diffusion strip package insert | Biomerieux amphotericin B Etest package insert (or similar) |
5.1
Vortex
5.2
Spectrophotometer or nephelometer
5.3
Incubator set to 35°C
6.1
Sabouraud’s Dextrose (SAB) agar plates (example: ThermoFisher Scientific, Product #R112562) or equivalent
6.2
RPMI-1640 media (with glutamic acid and phenol red, without bicarbonate; example: Sigma-Aldrich, Product #R1383) or equivalent
6.3
MOPS (3-[N-morpholino] propanesulfonic acid) buffer (example: Sigma-Aldrich, Product #M3183) or equivalent
6.4
D-(+)-glucose (example: Sigma Aldrich, Item# G8270)
6.5
Sterile water (example: Fisher Scientific, Catalog #15-230-162)
6.6
Sodium hydroxide (NAOH) solution, 1mol/L (example: Millipore Sigma, Product #1370311002) or equivalent
6.7
0.2 micron filter (example: Fisher Scientific, Catalog #03-377-137) or equivalent
6.8
BD Bacto Dehydrated Agar (example: VWR, Catalog #214010) or equivalent
6.9
Petri dish (100 x 15mm) (example: Fisher Scientific, Catalog #FB0875712)
6.10
Sterile glass tubes with screw top lid or cuvettes for use with your spectrometer or nephelometer – (make sure they fit your instrument)
6.11
Micropipet, size 10-100µL
6.12
Micropipet tips, size 10-100µL
6.13
Plastic box with lid for incubator to keep up humidity
6.14
Sterile cotton swabs (example: BD Falcon Polyester Fiber-tipped Applicator Swab, Ref 220690) or equivalent
6.15
Plastic transfer pipets (example: Fisherbrand, Cat # 13-711-20) or equivalent
6.16
Gradient Diffusion strips ( store at -20°C) (example: Amphotericin B Etest strips; BioMerieux, SKU 526348) or equivalent
6.17
Quality control isolates C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 which are available from the American Type Culture Collection
6.18
Plastic 1 µL inoculating loop (example: Thermo Scientific™ 253287)
6.19
0.5 McFarland Standard if using a nephelometer (example: Thermo Scientific™ R20410)
6.20
Forceps
6.21
RPMI agar and Sabouraud Dextrose agar plates are available from commercial sources. When necessary, RPMI agar may be prepared using the formulation in Section 7.0.
7.1
Dissolve 10.4 g RPMI 1640 powder (with glutamic acid and phenol red, without bicarbonate), 34.5 g MOPS (3-[N-morpholino] propanesulfonic acid) buffer, and 20g D- glucose into 450 ml of distilled H2O.
7.2
Adjust to pH 7.0 at 25°C using 1mol/L NaOH.
7.3
Filter sterilize using a 0.2 micron filter and place in hot water bath to increase temperature to 50°C.
7.4
Add 15 g of agar to 400 mL of distilled H2O.
7.5
Autoclave for 20 min to sterilize.
7.6
Add the sterile agar to the RPMI broth, adjust the final volume to 1 L with sterile distilled H2O and stir to mix.
7.7
In a BSC, aseptically dispense 25 mL per petri dish and allow the agar to solidify, with lid ajar. Once the dishes have come to room temperature, store at 4°C for up to 6 months.
8.1
Standard personal protective equipment should follow institutional guidelines but should consist of at least labcoat, gloves, and safety glasses which should be worn at all times.
8.2
Surfaces and supplies should be decontaminated using 1:128 Lysol solution or other decontaminant for yeasts approved by your institution.
8.3
Most Candida species can be processed at the bench using standard BSL2 precautions (lab coat, gloves, and safety glasses), if consistent with your institution’s safety procedures. One exception: Candida auris cultures must be handled in a BSC. Gloves should be worn at all times and BSC decontaminated with OxivirTB or freshly made 10% bleach.
9.1
Quality control isolates C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 should both be run each time susceptibility testing is performed. QC isolates are aliquoted and stored frozen. A new frozen aliquot is plated every two weeks. Repeated passage of the QC isolates will result in susceptibility results that are outside of the QC range.
9.2
The QC range for these isolates can be found in CLSI document M60. (See Appendix B below). If the QC isolates are out of range, all of the other results should be discarded.
10.1
Starting from a frozen yeast stock
10.1.1
Streak the test isolate and two control isolates (see note below) on separate SAB agar plates and allow them to grow for 24 hours at 35-37°C.
10.1.2
Re-streak from this plate onto a new SAB agar plate, streaking for isolation of colonies, and allow this second plate to grow for 24 hours at 35-37°C.
10.1.3
The original SAB agar plate containing growth directly from the freezer (step 10.1.1) may be kept at 4°C and used for up to 2 weeks. Discard this plate after 2 weeks.
10.2
Starting from water stock or an original plate
10.2.1
Streak the test isolate and two control isolates (see note below) for isolation on separate SAB agar plates and allow them to grow for 24 hours at 35-37°C. It is only necessary to subculture once.
NOTE: Be sure to also use a fresh culture of the two quality control isolates ATCC 22019 (C. parapsilosis) and ATCC 6258 (C. krusei).
11.1
Remove RPMI agar plates from 4°C storage and allow them to reach room temperature.
11.2
Take the appropriate number of gradient diffusion strips out of -20°C storage and allow them to come to room temperature.
Note: Do not touch the face of the strips, and do not lay down on a non-sterile surface. Always handle the strips by the end, and always use forceps.
11.3
If using a spectrophotometer, follow the instructions in the owners manual for use. Briefly turn on and allow to warm up for approximately 5-10 minutes. Once the machine is warmed up, choose percent (%) transmittance option. Set the wavelength to 530 nm.
Note: If using a nephelometer, it does not need to be warmed up.
11.4
Label one 13×100 glass tube (or whatever tube fits the instrument) per isolate with the isolate identifying number. Aliquot 2mL of sterile water into the glass tube. Place tube into the spectrophotometer and press the button labelled “0 ABS 100% T” to blank the machine to 0% transmittance.
11.5
Using the wooden end of a sterile wooden cotton swab, making sure not to touch or contaminate the wooden end (or you can use a sterile plastic 1 µL inoculating loop), pick 2-3 small yeast colonies (1 mm or smaller) from the fresh culture plate (prepared in step 10) and suspend them in the 2 ml of sterile water in the glass tube by tapping against the inside of the tube.
11.6
Carefully vortex the resulting suspension for 15 seconds to remove clumps.
11.7
Density reading of the yeast suspension using a spectrophotometer.
11.7.1
Place the tube in the spectrophotometer. The percent transmittance of light of the suspension should fall within 80% to 82%.
11.7.1.1
If percent transmittance of light is below 80%, add additional sterile water.
11.7.1.2
If percent transmittance is of light above 82%, add additional yeast cells.
11.7.2
When percent transmittance of light falls within 80% to 82%, the yeast stock suspension will yield 1-5 x 106 cells per ml.
11.8
Density reading of the yeast suspension using the nephelometer.
11.8.1
Place the tube in the nephelometer. The reading should fall within the 0.5 McFarland range.
11.8.1.1
If the density is too high, add additional sterile water.
11.8.1.2
If the density is too low, add additional yeast cells.
11.8.1.3
When the reading falls within the 0.5 McFarland range, the yeast stock suspension will yield 1-5 x 106 cells per ml.
11.9
Dip a sterile cotton swab in the glass tube with the adjusted yeast cell suspension, and remove excess fluid by pressing it against the inside wall of the tube.
11.10
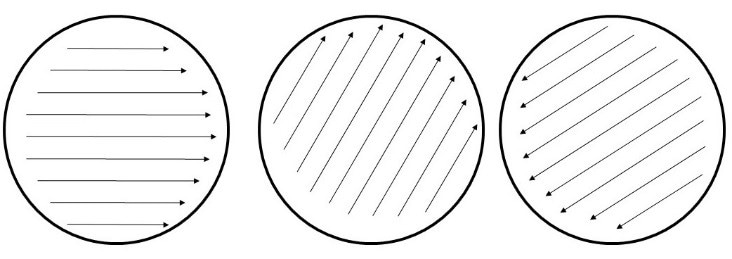
Streak an RPMI agar plate completely in three directions as shown below, making sure the entire plate is covered. Do not re-dip into the adjusted yeast cell suspension at any time (this will result in an inoculum that is too dense and can lead to false resistance). This is different from what the package insert suggests to do.
11.11
Allow the plate to air dry in the BSC with the lid cracked open for up to 15 minutes if it is excessively moist.
11.12
Pick up the gradient diffusion strip using forceps, being careful to only touch the strip upward of the dark black line.
NOTE: The area below this dark black line contains antifungal drug and should not be touched by hand or forceps.
11.13
Apply the gradient diffusion strip to plates with the MIC numbers facing upward, being careful to not allow bubbles under the strip.
Note: If bubbles occur, remove them by gently pressing down with the forceps, being careful not to slide the strip. DO NOT lift the strip and replace it.
11.14
Replace plate lid and place plate in a container that will maintain humidity (such as a plastic shoe box) and incubate at 35°C for 24 hours.
12.1
After 24 hours of growth at 35°C, the gradient diffusion strips are ready to read.
12.2
Gradient diffusion strips are read visually by opening the plate and observing how the growth intersects with the testing strip.
12.3
First, read the gradient strips for the two QC isolates. Using Appendix A, make sure the QC isolates are in range. If either of the isolates are not in range, the values for the antifungal in question should not be used.
12.4
If a sufficient lawn of growth is not obtained after 24 hours, the plate can be incubated for up to another 24 hours.
Note: Only in rare occasions and with certain species would this occur if the steps in Section 11 are performed correctly.
12.5
An ellipse where growth is not present may be seen in the lawn of growth, with the growth concentrated near the MIC endpoint. Isolates which are entirely resistant to that antifungal drug will show no ellipse.
12.6
Always read the value on the side of the strip with the higher of the MIC values. If growth is inhibited between two MIC values, use the greater of the two values as the MIC.
12.7
The interpretation of the MIC value is dependent upon the antifungal drug class. For polyenes (amphotericin B), the MIC is interpreted as the value where there is 100% growth inhibition. For azole and echinocandin antifungals, the MIC is interpreted as the value where there is 80% growth inhibition.
12.8
See Appendix B (Etest manufacturer’s instructions for interpretive guidelines).
13.1
Reference and alert values for resistant isolates can be found in CLSI document M60.
14.1
Clinical and Laboratory Standards Institute. 2017. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard – Fourth Edition, CLSI Document M27-A4. Clinical and Laboratory Standards Institute, Wayne, PA.
14.2
Clinical and Laboratory Standards Institute. 2017. Performance Standards for Antifungal Susceptibility Testing of Yeasts; Approved Standard – First Edition, CLSI Document M60. Clinical and Laboratory Standards Institute, Wayne, PA.
Appendix A
Quality Control Ranges
Recommended 24-hour Minimal Inhibitory Concentration Limits for Two Quality Control Strains for Broth Microdilution Procedures
Reference: Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antifungal Susceptibility Testing of Yeasts. 1st ed. CLSI supplement M60 (ISBN 1-56238- 828-2 [Print]; ISBN 1-56238-829-0 [Electronic]). Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA, 2017.
| Organism | Antifungal Agent | MIC Range, µg/mL | Mode | % of MICs Within Range |
|---|---|---|---|---|
| Candida parapsilosis ATCC®• 22019 | Amphotericin B | 0.25–2 | 0.5 | 97.1 |
| Anidulafungin | 0.25–2 | 1 | 95 | |
| Caspofungin | 0.25–1 | 0.5 | 96.7 | |
| Fluconazole | 0.5–4 | 2 | 98.2 | |
| Flucytosine | 0.06–0.25 | 0.12 | 99.2 | |
| Isavuconazole | 0.015–0.06 | 0.06 | 90.5 | |
| Itraconazole | 0.06–0.5 | 0.25 | 95.8 | |
| Ketoconazole | 0.03–0.25 | 0.06/0.12 | 97.5 | |
| Micafungin | 0.5–2 | 1 | 100 | |
| Posaconazole | 0.03–0.25 | 0.12 | 96.7 | |
| Voriconazole | 0.016–0.12 | 0.06 | 100 | |
| Candida krusei ATCC® 6258 | Amphotericin B | 0.5–2 | 1 | 100 |
| Anidulafungin | 0.03–0.12 | 0.06 | 97.9 | |
| Caspofungin | 0.12–1† | 0.5† | 98.8 | |
| Fluconazole | 8–64 | 16 | 100 | |
| Flucytosine | 4–16 | 8 | 97.5 | |
| Isavuconazole | 0.06–0.5 | 0.25 | 95.2 | |
| Itraconazole | 0.12–1 | 0.5 | 95.8 | |
| Ketoconazole | 0.12–1 | 0.5 | 95.4 | |
| Micafungin | 0.12–0.5 | 0.25 | 99.6 | |
| Posaconazole | 0.06–0.5 | 0.25 | 100 | |
| Voriconazole | 0.06–0.5 | 0.25 | 98.3 |
Appendix B
Etest Antifungal Reading Guide from the package insert for Etests from Biomerieux [PDF – 2 pages]. Edition 2013/02.
Azoles
When trailing endpoints occur, read the MIC at the first point of significant inhibition of growth i.e. the so-called 80% inhibition as judged by the naked eye. When trailing is strong, especially with species such as C. glabrata with reduced azole susceptibility, and for smaller ellipses at higher MIC values, read the plate against a strong light source to facilitate visual detection of the endpoint at 80% inhibition.
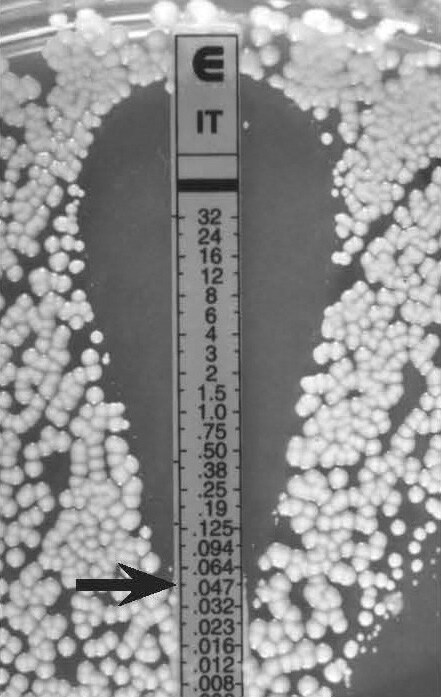
Figure 7.
C. albicans, clear endpoint
MIC 0.047 µg/ml

Figure 8.
Candida spp., slim ellipse with trailing colonies
MIC: 0.094 µg/ml
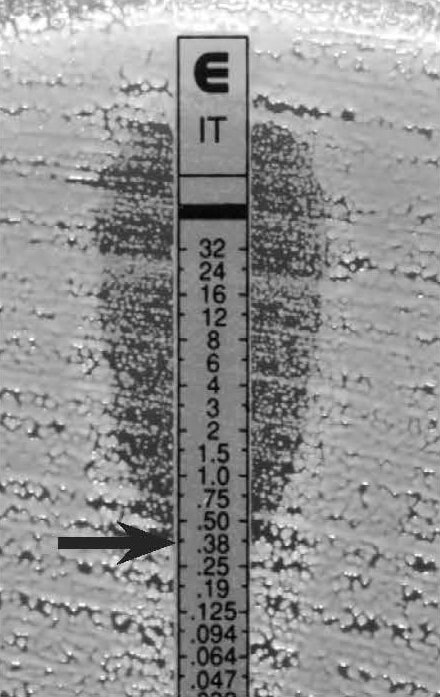
Figure 9.
C. tropicalis, discernable ellipse with a lawn of microcolonies
MIC 0.38 µg/ml
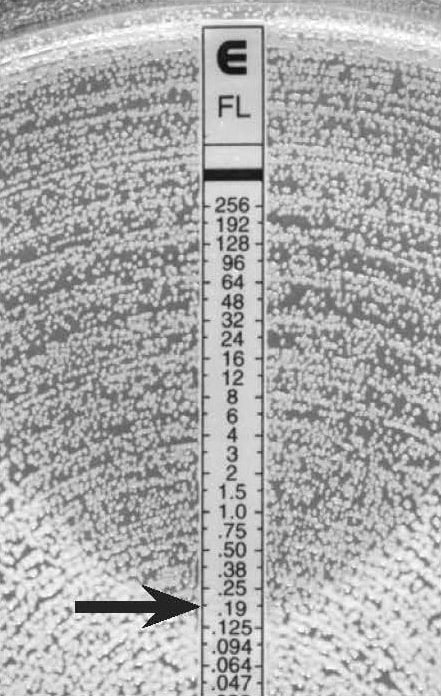
Figure 10.
C. albicans, discernable ellipse with a lawn of microcolonies
MIC: 0.19 µg/mL

Figure 11.
C. parapsilosis, trailing colonies
MIC: 0.25 µg/mL
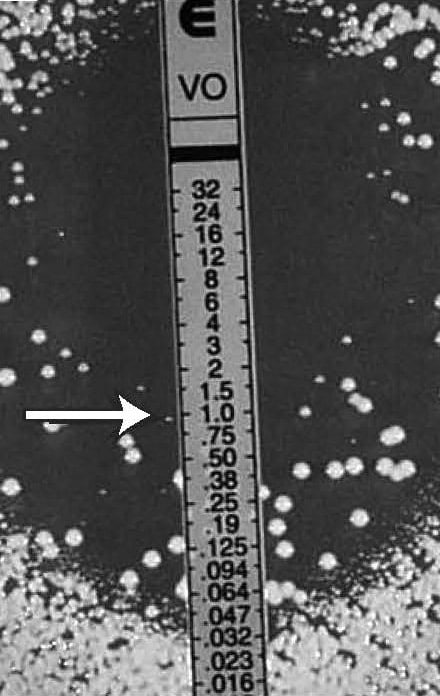
Figure 12.
Candida spp., macrocolonies in the ellipse, (potential heteroresistance)
MIC: 1 µg/mL

Figure 13.
C. krusei, macrocolonies in the ellipse
MIC: ≥256 µg/mL
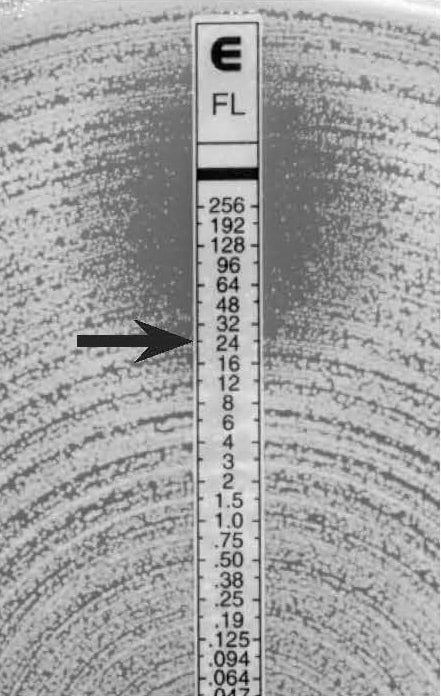
Figure 14.
C. glabrata, higher MIC with trailing colonies
MIC: 24 µg/mL

Figure 15.
Candida spp., trailing colonies
MIC: 0.094 µg/mL
Amphotericin B
Read endpoint at complete inhibition of growth i.e. 100% inhibition including all microcolonies, hazes and isolated colonies.

Figure 16.
Candida spp., clear endpoint
MIC 0.094 µg/mL

Figure 17.
Candida spp., macrocolonies in the ellipse
MIC: 3 µg/mL
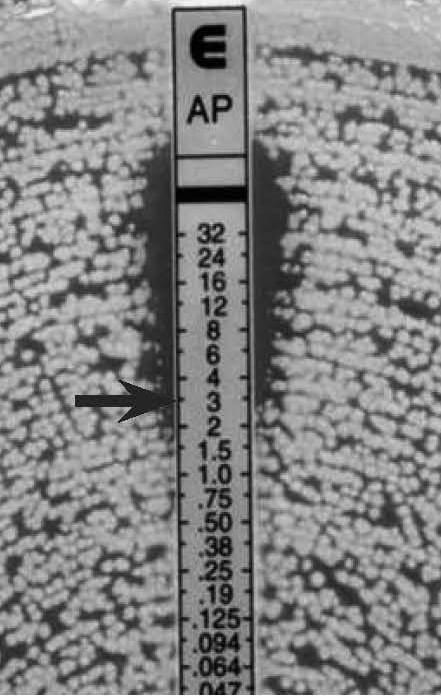
Figure 18.
Candida spp., slim ellipse and microcolonies
MIC: 3 µg/mL
Echinocandins
When trailing endpoints or other growth/inhibition phenomena occur, read the MIC at the first visual point of significant inhibition i.e. 80% inhibition.
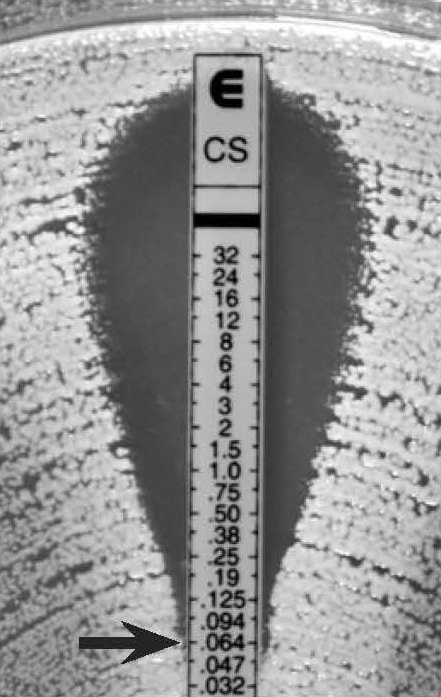
Figure 22.
C. albicans “dip effect” – read the MIC at the bottom of the dip
MIC: 0.064 µg/mL

Figure 23.
C. tropicalis, ellipse with regrowth at the higher concentration range – ignore re-growth
MIC: 0.125 µg/mL
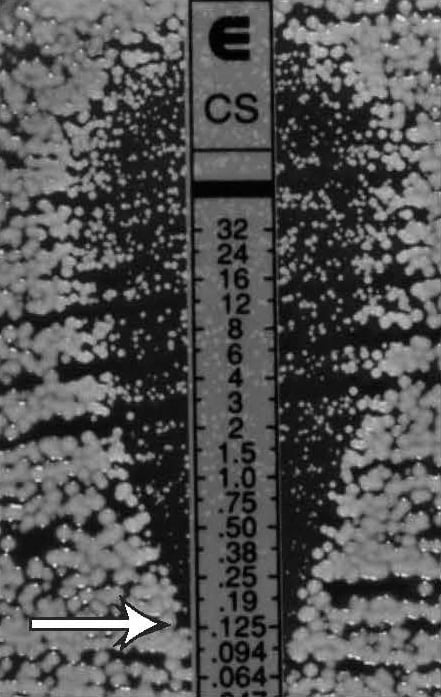
Figure 24.
Candida spp., discernable ellipse with microcolonies
MIC: 0.125 µg/mL
DISCLAIMER:
The Mycotic Diseases Branch laboratory developed this document as an example test procedure for Antifungal Susceptibility Testing by Gradient Diffusion. It is the responsibility of the testing laboratory to ensure content and format are modified as necessary to meet applicable regulatory requirements, quality management system standards, and chemical, radiological and biological safety requirements. This is not a controlled document, and the described test methods are subject to change without notice. It is the responsibility of the testing laboratory to ensure the information within this document remains applicable. Contact the test developer at gyi2@cdc.gov to find out whether any changes have been made.
Use of trade names and commercial sources is for identification only and does not constitute endorsement by the Public Health Service or by the United States Department of Health and Human Services.