Chapter 22: Laboratory Support for Surveillance of Vaccine-Preventable Diseases
Authors: Sandra Roush, MT, MPH; Bernard Beall, PhD; Lesley McGee, PhD; Michael Bowen, PhD; Steven Oberste, PhD; Paul Rota, PhD; Maria Lucia Tondella, PhD; Annemarie Wasley, ScD; Xin Wang, PhD; Brian Wakeman, PhD; Rachel Suzanne Beard, PhD
Surveillance of Vaccine-Preventable Diseases
Surveillance for vaccine-preventable diseases (VPDs) requires close collaboration with clinicians, public health professionals, and laboratory scientists. Public health surveillance relies on both clinical and laboratory reports of VPDs; therefore, appropriate specimen collection, transport, and laboratory testing are essential. This chapter provides guidelines on specimen collection and interpretation of laboratory results for each VPD.
Each public health professional diagnosing and treating vaccine-preventable diseases should identify laboratory support for his or her clinical and public health practice. Table 1 lists appropriate tests for VPDs and provides names and contact information for laboratories and laboratory personnel. In addition to the guidelines presented in this chapter, state health department personnel can provide additional guidance on specimen collection, transport, and related information.
Table 1. Contact persons for VPD surveillance laboratory support
| Disease | Test name | Lab name | Lab contact/Phone |
|---|---|---|---|
| Diphtheria | Culture
Toxigenicity testing PCR |
CDC Pertussis and Diphtheria Laboratory | Contact the Meningitis and Vaccine Preventable Diseases Branch to be directed to the appropriate staff
404-639-3158 |
| Haemophilus influenzae | Culture
Real-time PCR Serotyping slide agglutination Molecular typing (WGS) |
CDC Bacterial Meningitis Laboratory | Dr. Henju Marjuki vsd1@cdc.gov 404-639-2803 |
| Hepatitis A | Laboratory Branch | Dr. Saleem Kamili sek6@cdc.gov 404-639-4431 |
|
| Hepatitis B | Laboratory Branch | Dr. Saleem Kamili sek6@cdc.gov 404-639-4431 |
|
| Influenza | Culture/viral isolation
Antigen detection RT-PCR/ RT-PCR |
Influenza Division, Virology, Surveillance, and Diagnosis Branch | John Barnes Genomics & Diagnostics Team Lead fzq9@cdc.gov Email: flusupport@cdc.gov 404-639-1587 |
| Measles | IgM antibody
IgG antibody Virus isolation RT-PCR Viral genotyping |
Viral Vaccine-Preventable Diseases Branch | Dr. Paul Rota par1@cdc.gov 404-639-4181 |
| Meningococcal disease | Culture
Real-time PCR Serogrouping slide agglutination |
CDC Bacterial Meningitis Laboratory | Dr. Henju Marjuki vsd1@cdc.gov 404-639-2803 |
| Mumps | IgM antibody IgG antibody Virus isolation RT-PCR Viral genotyping |
Vaccine Viral Preventable Diseases Branch | Dr. Stephen Crooke SCrooke@cdc.gov 404-718-4003 |
| Pertussis | Culture
PCR Serology |
CDC Pertussis and Diphtheria Laboratory | Dr. Lucia Tondella mlt5@cdc.gov 404-639-1239 |
| Pneumococcal disease | Culture
PCR Susceptibility testing Serotyping (conventional or PCR-based) Genotyping Antibiotic resistance |
CDC Streptococcus Laboratory | Dr. Lesley McGee LMcGee@cdc.gov 404-639-0455 or |
| Poliomyelitis | RT-PCR Sequencing |
CDC Polio/Picornavirus Laboratory | PicornaLab@cdc.gov |
| Rotavirus | Antigen EIA
RT-PCR, qRT-PCR Genotyping Sequencing |
CDC Rotavirus Laboratory | Rashi Gautam hsr7@cdc.gov 404-639-1628 |
| Rubella | IgM antibody
IgG antibody IgG Avidity Culture RT-PCR |
Vaccine Viral Preventable Diseases Branch | Dr. Ludmila Perelygina LPerelygina@cdc.gov 404-639-4812 |
| Congenital rubella syndrome | IgM antibody
IgG antibody Culture RT-PCR Serology |
Vaccine Viral Preventable Diseases Branch | |
| Varicella | PCR
Serology DFA* Viral Culture* |
Viral Vaccine-Preventable Diseases Branch | Dr. R. Suzanne Beard rbeard@cdc.gov 404-639-5455 |
General Guidelines for Specimen Collection and Laboratory Testing
Specimen collection and shipping are important steps in obtaining laboratory diagnosis for VPDs. Guidelines have been published for specimen collection and handling for viral and microbiologic agents. Information is also available on using Centers for Disease Control and Prevention (CDC) laboratories as support for reference and disease surveillance, which includes:
- a central website for requesting lab testing.
- the form required for submitting specimens to CDC (Appendix 23, CDC Form #50.34 [2.74 MB, 2 pages];
- information on general requirements for shipment of etiologic agents (Appendix 24 [4 pages])—although written to guide specimen submission to CDC, this information may be applicable to the submission of specimens to other laboratories; and
- the CDC Infectious Diseases Laboratories Test Directory that has a list of available tests, detailed information on appropriate specimen types, collection methods, specimen volume, and points of contact.
In addition, there are 4 VPD Reference Centers [2 pages]—public health laboratories that perform testing for 7 VPDs using standardized methods developed by CDC.
Disease-specific Guidelines for Specimen Collection and Laboratory Testing
This chapter provides a quick reference summary of the laboratory information in relation to Vaccine-Preventable Diseases from Chapters 1-17 of this manual. Confirmatory and other useful tests for surveillance of vaccine-preventable diseases are listed below in Table 2.
Table 2. Confirmatory and other useful tests for the surveillance of vaccine-preventable diseases
| Disease | Confirmatory tests | Other useful tests |
|---|---|---|
| Diphtheria | Culture Toxigenicity testing |
PCR
MALDI-TOF |
| Haemophilus influenzae | Culture
Real-time PCR |
Serotyping slide agglutination or PCR (identification of capsular type of encapsulated strains) Antigen detection Molecular typing |
| Hepatitis A | IgM anti-HAV (positive) | Total anti-HAV;
IgG anti-HAV (markers of immunity) |
| Hepatitis B | IgM anti-HBc (acute infection) HBsAg (acute or chronic infection)* |
Anti-HBs (marker of immunity) Total anti-HBc (marker of past or present infection) PCR for HBV DNA (marker of current infection) |
| Influenza | Culture Antigen detection (EIA, IFA, EM) RT-PCR |
|
| Measles | IgM RT-PCR Virus isolation |
IgG IgG for seroconversion or 4-fold titer rise Avidity (case classification) |
| Meningococcal disease | Culture | Serogrouping
Slide agglutination or PCR Antigen detection Molecular typing |
| Mumps | Virus isolation
RT-PCR |
IgG for seroconversion or 4-fold titer rise (not recommended) |
| Pertussis | Culture PCR |
Serology |
| Pneumococcal disease | Culture PCR |
WGS-based deduction of all strain features (serotype, antimicrobial resistance, MLST genotype
|
| Poliomyelitis | Sequencing | |
| Rotavirus | RT-PCR/qRT-PCR | |
| Rubella | Paired sera for IgG IgM |
Culture |
| Tetanus | There are no lab findings characteristic of tetanus | Serology (for immunity testing) |
| Varicella | PCR
Culture** |
DFA
Serology Genotyping |
Abbreviations:EIA, enzyme-linked immunosorbent assay; IFA, indirect fluorescent antibody; EM, electron microscopy; PCR, polymerase chain reaction; MLST, multilocus sequence typing; PFGE, pulsed field gel electrophoresis; CSF, cerebral spinal fluid; IgG, immunoglobulin G; IgM, immunoglobulin M; HAV, hepatitis A virus; HBV, hepatitis B virus; anti-HBc; hepatitis B core antibody; HBsAg, hepatitis B surface Ag; qRT-PCR, quantitative reverse transcription polymerase chain reaction; PCR, polymerase chain reaction.
* Confirmation of HBsAg positive results by HBsAg neutralization assay should be performed as specified in test package insert.
**PCR is the recommended gold standard test but culture can be performed.
Table 3 summarizes specimen collection procedures for laboratory testing. Because some specimens require different handling procedures, it is essential to check with the diagnostic laboratory prior to shipping. When in doubt about what specimens to collect, timing of specimen collection, or where or how to transport specimens, call the state health department and the state laboratory or the CDC.
Table 3. Specimen collection for laboratory testing for VPDs
| Disease | Test name | Specimens to take | Timing for specimen collection | Transport requirements | Collection requirements |
|---|---|---|---|---|---|
| Diphtheria | Culture
Note: ALERT lab that diphtheria is suspected, so that tellurite-containing media will be used. |
Swab of nose, throat, membrane | ASAP, when diphtheria is suspected | <24 hrs: Amies or similar transport medium
≥24 hrs: silica gel sachets |
State health departments may call CDC diphtheria lab at 404-639-1231 or 404-639-1239. |
| Diphtheria | PCR
Note: ALERT lab that diphtheria is suspected, so that specific PCR assay will be used. |
Swabs (as above), pieces of membrane, biopsy tissue | Take these specimens at same time as those for culture | Swabs, silica gel sachet,
or a sterile dry container at 4°C |
State health departments may call CDC diphtheria lab at
404-639-1231 or 404-639-1239. |
| Diphtheria | Toxigenicity testing (Elek test) | Isolate from culture (above) | After C. diphtheriae has been isolated | Transport medium, such as Amies medium, or silica gel sachets | State health departments may call CDC diphtheria lab at 404-639-1231 or 404-639-1239. |
| Diphtheria | Serology (antibodies to diphtheria toxin) Note: Collect paired sera, taken 2-3 weeks apart.This test is currently not available at CDC. |
Serum | Before administration of antitoxin | Frozen (-20°C) | Not useful if diphtheria antitoxin was administered. |
| Haemophilus influenzae type b | Culture Note: Request lab to conduct sero-typing on any Hi isolate from any normally sterile site. |
Blood | ASAP | Blood culture bottles w/broth
or lysis-centrifugation tube |
Collect 3 separate samples in a 24-hr period. |
| Haemophilus influenzae type b | Culture
Other normally sterile site |
CSF | ASAP | Sterile, screw-capped tube | |
| Haemophilus influenzae type b | Serotyping slide agglutination | Culture isolate | Chocolate slant, frozen, lyophilized
or silica gel pack |
Highest priority are isolates from persons <15 years. | |
| Haemophilus influenzae type b | Antigen detection | CSF | ASAP | Sent frozen on blue ice packs | |
| Haemophilus influenzae type b | PCR for identification and serotyping | Any normally sterile site | ASAP | Sent frozen on blue ice packs | |
| Hepatitis A | IgM anti-HAV | Serum | ASAP after symptom onset (detectable up to 6 months) | All sera to be tested for serologic markers of HAV and HBV infection can be kept at ambient temperatures, refrigerated (<48 hours) for short term. For longer than 48 hours storage, sera should be frozen. | Follow standard procedures for serum separation. |
| Hepatitis A | Total anti-HAV
Note: Measures both IgM and IgG. IgG anti-HAV |
Serum | No time limit | Samples can be kept at ambient temperatures, refrigerated (<48 hours) for short term. For longer than 48 hours storage, sera should be frozen. | Follow standard procedures for serum separation. |
| Hepatitis A | HAV RNA by PCR | Serum | ASAP after symptom onset (detectable up to 2 weeks) | Store and ship samples frozen | Follow standard procedures for serum separation. |
| Hepatitis B | IgM anti-HBc | Serum | ASAP after symptom onset (detectable up to 6 months) | Samples can be kept at ambient temperatures, refrigerated (<48 hours) for short term. For storage longer than 48 hours, sera should be frozen. | Follow standard procedures for serum separation. |
| Hepatitis B | HBsAg
Note: HBsAg-positive results should be confirmed by HBsAg neutralization assay as specified in the package insert for each assay. |
Serum | ASAP after symptom onset | Samples can be kept at ambient temperatures, refrigerated (<48 hours) for short term. For longer than 48 hours storage, sera should be frozen. | Follow standard procedures for serum separation. |
| Hepatitis B | Anti-HBs | Serum | 1–2 months after vaccination | Samples can be kept at ambient temperatures, refrigerated (<48 hours) for short term. For longer than 48 hours storage, sera should be frozen. | Follow standard procedures for serum separation. |
| Influenza | Culture/viral isolation | Nasal wash, nasopharyngeal aspirates, nasal/throat swabs, transtracheal aspirate, bronchoalveolar lavage | Within 72 hours of onset of illness | Transport specimens at 4°C if tests are to be performed within 72 hours; otherwise, freeze at -70°C until tests can be performed. | |
| Influenza | Antigen detection and RT-PCR | Nasal wash, nasopharyngeal aspirate, nasal/throat swabs, gargling fluid, transtracheal aspirates, bronchoalveolar lavage | Within 72 hours of onset of illness | Transport specimens at 4°C if tests are to be performed within 72 hours; otherwise, freeze at -70°C until tests can be performed. | Note: Save an aliquot of the clinical sample for confirmation and isolation. Viral isolates may be further characterized by WHO/CDC. |
| Measles | Virus isolation
RT-PCR |
Nasopharyngeal aspirates, throat swabs, urine, heparinized blood | Collect at same time as samples for serology (best within 3 days of rash onset) | Transport specimens at 4°C if tests are to be performed within 72 hours; otherwise, freeze at -70°C until tests can be performed. | Note: PCR for genotyping.Collect up to 10 days from rash onset. |
| Measles | IgM antibody | Serum | ASAP after rash onset and repeat 72 hours after onset if first negative | Ship on cold pack | Note: IgM is detectable for at least 30 days after rash onset. |
| Measles | IgG antibody | Paired sera | Acute: ASAP after rash onset (7 days at the latest) Convalescent: 14-30 days after acute |
||
| Meningococcal disease | Culture**
Note: Request that lab conduct serogrouping on any N. meningitidis isolate |
Blood | ASAP | Blood culture bottles w/broth
or lysis-centrifugation tube |
|
| Meningococcal disease | Culture**
Note: Request that lab conduct serogrouping on any N. meningitidis isolate |
CSF
Other normally sterile site |
ASAP | Sterile, screw-capped tube | |
| Meningococcal disease | Serogrouping slide agglutination | Isolate from culture (above) | Slant, frozen, lyophilized
or silica gel pack. |
||
| Meningococcal disease | PCR for detection and serogrouping | Any normally sterile site | ASAP | Sent frozen on blue ice packs | |
| Meningococcal disease | Antigen detection | Any normally sterile site | ASAP | Sent frozen on blue ice packs | |
| Mumps | Virus isolation
RT-PCR |
Buccal*/parotid swabs; CSF only for aseptic meningitis; urine for cases of orchitis | Ideally 0-3 days after parotitis onset but up to 10 days | Transport specimens at 4°C if tests are to be performed within 72 hours; otherwise, freeze at -70°C until tests can be performed. | Massage the salivary/parotid gland area for 30 seconds prior to swab collection. |
| Mumps | IgM antibody | Serum | Ideally 3 days after parotitis onset | Ship frozen on dry ice | |
| Mumps | IgG antibody | Paired sera for seroconversion
or 4-fold rise in titer |
Acute: ideally 0–3 days after parotitis onset Convalescent: 2 weeks after acute | Ship frozen on dry ice | Testing four-fold rise in titer not recommended for previously vaccinated persons. |
| Pertussis | Culture
Note: Inoculate selective primary isolation media such as charcoal horse blood agar or Bordet-Gengou as soon as possible. A negative culture does NOT rule out pertussis. |
Posterior nasopharyngeal swab or aspirate | Within the first 2 weeks of cough onset | Swabs: half-strength charcoal horse blood agar at 4°C
Swabs in Regan-Lowe transport Aspirates: in saline in capped syringe at 4°C |
Use polyester (such as Dacron), rayon, or nylon nasopharyngeal swab. Flocked swabs are preferred. Shaft may be flexible plastic, aluminum, or twisted wire. Aspirates may be collected with a syringe and catheter by introducing a small amount of saline into the nasopharyngeal cavity and collecting it in the syringe. |
| Pertussis | PCR
Note: PCR should be validated with culture when possible. |
Nasopharyngeal swab or aspirate | Within the first 2 weeks of cough onset | Swabs in Regan-Lowe transport at 4◦C
Aspirates in saline in capped syringe at 4◦C For PCR testing only, swab may be placed in a sterile tube and sent at 4°C (short term storage) or -20°C or below (long-term storage) |
Use polyester (such as Dacron) rayon, or nylon pharyngeal swab. Flocked swabs are preferred. Shaft may be flexible plastic, aluminum, or twisted wire. Aspirates may be collected with a syringe and catheter by introducing a small amount of saline into the nasopharyngeal cavity and collecting it in the syringe. |
| Pertussis | Serology | Serum | 2 to 8 weeks of cough onset | Short term at 4°C; long term at -20°C or below | Serologic results are currently not accepted as laboratory confirmation for purposes of national surveillance. |
| Pneumococcal disease | Culture | Normally sterile site | As soon as possible after onset of clinical illness but before administration of antibiotics | Blood culture bottles w/broth or lysis-centrifugation tube or, if from another sterile site, a sterile, screw-capped tube | Collect 2 separate blood samples in a 24-hr period. Most other sterile specimens (e.g., CSF) are collected only once. |
| Pneumococcal disease | PCR | Normally sterile site | ASAP, soon after administration of antibiotics is a viable option | Send specimen frozen on blue ice packs | PCR |
| Pneumococcal disease | PCR deduction of serotype | Culture-negative sterile site specimen | Specimen frozen immediately | PCR deduction of serotype | |
| Pneumococcal disease | Susceptibility testing | Pure culture | Slant, frozen, or silica packet | Susceptibility testing | |
| Pneumococcal disease | Serotyping | Pure culture | Slant, frozen, or silica packet | Serotyping | |
| Poliomyelitis | Culture | Stool, pharyngeal swab, CSF | Acute | Sterile, screw-capped container | No carrier for stool; saline buffer for swabs
Note: Maintain frozen or transport rapidly to lab; avoid desiccation of swab specimens. |
| Poliomyelitis | Intratypic differentiation | Isolate from culture (above) | Note: Maintain frozen or transport rapidly to lab; avoid desiccation of swab specimens. | ||
| Poliomyelitis | Serology | Paired sera | Acute: ASAP
Convalescent: 3 weeks after acute |
||
| Rotavirus gastroenteritis | EIA, RT-PCR, qRT-PCR, genotyping, sequencing | Stool | First to fourth day of illness optimal | Sterile, screw-capped container | Bulk stool
Keep frozen or transport rapidly to lab on cold packs; avoid multiple freeze-thaw cycles. |
| Rotavirus-associated seizures | RT-PCR, qRT-PCR | CSF | ASAP after symptoms begin | Sterile, screw-capped container | No carrier.
Keep frozen; avoid multiple freeze-thaw cycles. |
| Rubella | IgM antibody | Serum | ASAP, and repeat 96 hours after onset if first negative | Maintain at 4°C and ship on ice |
|
| Rubella | IgG antibody | Serum
Paired sera |
Acute: ASAP after rash onset (7 days at the latest)
Convalescent: 14–30 days after acute |
Maintain at 4°C and ship on ice | Paired sera must be run in parallel |
| Rubella | Culture/PCR | Nasopharyngeal swab/wash, throat swab, urine. | Collect at same time as samples for serology (best within 3 days of rash onset and no later than 10 days post onset) | Viral transport media; ship frozen or on ice | Note: Maintain frozen (except urine) or transport rapidly to lab; avoid desiccation of swab specimens. |
| Congenital rubella syndrome (CRS) | IgM antibody | Serum | As soon as possible, within 6 months of birth | Maintain at 4°C and ship on ice | |
| Congenital rubella syndrome (CRS) | IgG antibody | Serum | After 9 months of age, but before vaccination with MMR vaccine | Maintain at 4°C and ship on ice | Note: Confirmation is by documenting persistence of serum IgG titer beyond the time expected from passive transfer of maternal IgG antibody. |
| Congenital rubella syndrome (CRS) | Culture/PCR | Nasopharyngeal swab/wash, urine, blood, cataracts | As soon as possible for confirmation; to monitor shedding in positive cases; after 3 months, every month until cultures are repeatedly negative | Viral transport media; ship frozen or on ice | Note: Maintain frozen (except urine) or transport rapidly to lab; avoid desiccation of swab specimens. |
| Varicella | Serology | Serum
CSF |
Immune status: collect after acute illness (3 weeks or more after symptom onset)
Diagnosis: paired sera, acute within 7–10 days of onset, convalescent 2–3 weeks after acute† |
Ship frozen at -20°C on dry ice | Refrigerate at 2°–8°C within 1 hour of collection, freeze -20°C within 48 hours |
| Varicella | VZV PCR | Material from skin lesions (papule, macule, or vesicles)
Scabs CSF |
Acute illness 2–3 days after rash onset | Ship scabs and dry swabs at RT overnight within 30 days of collection
Ship CSF or swabs in VTM frozen at -20°C overnight within 60 days of collection |
Scabs should be unroofed directly into a breakage resistant tube
Swabs of skin lesions may be placed directly into a storage tube or can be placed in VTM Scabs and dry Swab stored at RT Swabs in VTM Refrigerate at 2-8°C within 1 hour of collection up to 7 days, freeze -20°C up to 60 days CSF refrigerate immediately 2-8°C up to 14 days, freeze -20°C up to 60 days |
| Varicella | VZV PCR with Vaccine wild-type discrimination | Material from skin lesions (papule, macule, or vesicles)
Scabs CSF |
From day of rash onset until crusted lesions resolve | Ship scabs and dry swabs at RT overnight within 30 days of collection
Ship CSF or swabs in VTM frozen at -20°C overnight within 60 days of collection |
Scabs should be unroofed directly into a breakage resistant tube.
Swabs of skin lesions may be placed directly into a storage tube or can be placed in VTM. Scabs and dry swab stored at RT Swabs in VTM: Refrigerate at 2-8°C within 1 hour of collection up to 7 days, freeze -20°C up to 60 days CSF: refrigerate immediately 2-8°C up to 14 days, freeze -2°C up to 60 days |
** Neisseria meningitidis culture cannot be performed on specimens sent to CDC, but CDC is available to provide advice and answer questions on culture methods.
†Paired IgG acute- and convalescent-phase antibody tests are not practical for immediate clinical management. In vaccinated persons, a 4-fold rise may not occur.
A. Diphtheria (see Chapter 1)
Diagnostic tests used to confirm infection include isolation of Corynebacterium diphtheriae by culture and Elek testing of isolates for diphtheria toxin production.(1-3) The CDC Pertussis and Diphtheria Laboratory routinely performs culture to confirm C. diphtheriae and is currently the only laboratory in the United States that tests for toxin production. Although no other tests for confirming diphtheria are commercially available, CDC can perform a polymerase chain reaction (PCR) test on clinical specimens to confirm infection with a potentially toxigenic strain.(4) PCR can detect nonviable C. diphtheriae organisms from specimens taken after antibiotic therapy has been initiated.
PCR for species identification and detection of the diphtheria toxin gene, as performed by the CDC Pertussis and Diphtheria Laboratory, and MALDI-TOF provide supportive evidence for the diagnosis but do not confirm toxin production. These tests, when used on an isolate, should always be combined with a test that confirms toxin production, such as the Elek test.
If diagnosis or case classification is not straightforward based on available laboratory testing, CDC recommends consultation with its diphtheria subject matter experts for further discussion.
Isolation of C. diphtheriae by culture
Isolation of C. diphtheriae by bacteriological culture is essential for confirming diphtheria. The following should be considered:
- A clinical specimen for culture should be obtained as soon as possible when diphtheria (involving any site) is suspected, even if treatment with antibiotics has already begun.
- Specimens should be taken from the site of diphtheria infection, nose, throat, and, if present, from the diphtheritic membrane. If possible, swabs also should be taken from beneath the membrane.
- Other pathogens can cause a membrane in the throat and over the tonsils, including the pathogens listed below. The patient’s healthcare provider should be encouraged to perform laboratory tests, when appropriate, to rule out these conditions and organisms, which include
- Streptococcus ,
- Epstein-Barr virus and cytomegalovirus (both of which cause infectious mononucleosis syndrome),
- Arcanobacter haemolyticum,
- Candida albicans, and
- fusiform bacteria (which can cause Vincent’s angina).
- The laboratory should be alerted to the suspicion of diphtheria because identification of C. diphtheriae is aided by use of special culture media containing tellurite.
- Specimens from the nose and throat (i.e., both a nasal and an oropharyngeal swab) for culture should be obtained from all patients with suspected diphtheria and their close contacts.
- Isolation of C. diphtheriae from close contacts may confirm the diagnosis of the case, even if the patient’s culture is negative.
Biotype testing
After C. diphtheriae has been isolated, the biotype (substrain) should be determined. The 4 biotypes are gravis, mitis, intermedius, and belfanti.
Toxigenicity testing
In addition to determining biotype, toxigenicity testing using the Elek test should be performed to determine if the C. diphtheriae isolate produces toxin. These tests are not readily available in many clinical microbiology laboratories; isolates should be sent to a reference laboratory proficient in performing the tests.
Polymerase chain reaction testing
Clinical specimens collected for culture testing can also be used for PCR testing at CDC, if indicated. Because isolation of C. diphtheriae is not always possible (many patients have already received several days of antibiotics by the time a diphtheria diagnosis is considered), PCR can provide additional supportive evidence for the diagnosis of diphtheria. The PCR assay allows for detection of the diphtheria toxin gene (tox), identification of C. diphtheria through detection of an RNA polymerase gene (rpoB), and detection of C. ulcerans and C. pseudotuberculosis, also with a rpoB target. However, C. ulcerans and C. pseudotuberculosis are not differentiated from each other in this assay.
MALDI-TOF
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) is an additional technology (5) that can be used to rapidly identify bacterial species such as C. diphtheriae. The technique requires an isolate to identify the protein composition of microbial cells. However, this form of testing only confirms the bacterial species, does not confirm diphtheria toxin production, and is not available at the CDC Pertussis and Diphtheria Laboratory.
Serologic testing
Measurement of the patient’s serum antibodies to diphtheria toxin before administration of antitoxin may help in assessing the probability of the diagnosis of diphtheria. High antibody levels may indicate protection against diphtheria, and C. diphtheriae infection is less likely to produce a serious illness. However, if antibody levels are low, diphtheria cannot be accurately ruled out. The state health department or CDC can provide information on laboratories that offer this test; few laboratories have the capability to accurately test antibody levels.
Submission of C. diphtheriae isolates and clinical specimens
All isolates of C. diphtheriae from any anatomical site should be sent to the CDC Pertussis and Diphtheria Laboratory for reference testing. Clinical specimens from patients with suspected diphtheria for whom diphtheria antitoxin has been released for treatment should also be sent to the CDC Pertussis and Diphtheria Laboratory for culture, PCR, and toxigenicity testing. While not reportable, if C. ulcerans or C. pseudotuberculosis is identified, jurisdictions are asked to submit available specimens or isolates to the CDC Pertussis and Diphtheria Laboratory for further characterization.
Clinical specimens (swabs, pieces of membrane, heart biopsy tissue) can be transported to CDC following the current guidance in the CDC test order directory. For detailed information on specimen collection and shipping and to arrange for PCR testing, the state health department may contact the Meningitis and Vaccine Preventable Diseases Branch at 404-639-3158 to be directed to the appropriate staff.
B. Haemophilus influenzae type b (Hib) invasive disease (see Chapter 2)
Presumptive identification by Gram stain or antigen detection
The Gram stain is an empirical method for differentiating bacterial species into 2 large groups based on the chemical and physical properties of their cell walls. Gram-positive bacteria retain the primary stain while gram-negative bacteria take the color of the counterstain. A Gram stain can also serve to assess the quality of a clinical specimen. The Gram stain is useful for preliminary identification of likely H. influenzae, though is not a confirmatory test and cannot distinguish among H. influenzae type b (Hib) serotypes.
Because the type b capsular antigen can be detected in body fluids, including urine, blood, and cerebrospinal fluid (CSF) of patients, clinicians often request a rapid antigen detection test for diagnosis of Hib disease. Antigen detection may be used as an adjunct to culture or PCR, particularly in the diagnosis of patients who have received antimicrobial agents before specimens are obtained for culture. The method for antigen detection is latex agglutination, which is a rapid and sensitive method used to detect Hib capsular polysaccharide antigen in CSF; however, false-negative and false-positive reactions can occur.
If the Hib antigen is detected in CSF from a patient with meningitis but a positive result is not obtained from culture of sterile site, the patient should be considered as having a probable case of Hib disease and reported as such. Because antigen detection tests can be positive in urine and serum of persons without invasive Hib disease, persons who are identified exclusively by positive antigen tests in urine or serum should not be reported as cases.
Confirmation by culture and PCR
Confirming a case of Hib disease requires isolating H. influenzae or detecting H. influenzae DNA from a normally sterile body site. Normally, sterile sites for isolation of invasive H. influenzae typically include CSF, blood, joint fluid, pleural effusion, pericardial effusion, peritoneal fluid, subcutaneous tissue fluid, placenta, and amniotic fluid. Most hospital and commercial microbiologic laboratories have the ability to isolate H. influenzae. Isolates of H. influenzae are recommended to be tested for antimicrobial susceptibility to ampicillin, oneof the third-generation cephalosporins, chloramphenicol, and meropenem. Further antimicrobial susceptibility testing should be considered for isolates obtained from cases in which a treatment or chemoprophylaxis failure is suspected or in an outbreak setting.
Although culture is the gold standard for confirming H. influenzae, real-time PCR is an accepted alternative. In recent years, significant improvements have been made in both the sensitivity and specificity of PCR assays used for the detection of H. influenzae. Real-time PCR assays are available to detect DNA of H. influenzae and all 6 serotypes in blood, CSF, or other clinical specimens. A major advantage of PCR is that it allows for detection of H. influenzae from clinical samples in which the organism could not be detected by culture methods, such as when a patient has been treated with antibiotics before a clinical specimen is obtained for culture. Even when the organisms are nonviable following antimicrobial treatment, PCR can still detect H. influenzae DNA. Isolation of the bacterium is needed to test for antimicrobial susceptibility.
Several commercial multiplex PCR assays capable of simultaneously testing a single specimen for an array of pathogens that cause blood infections or meningitis/encephalitis are now available, primarily for clinical settings (e.g., FilmArray Blood Culture Identification Panel and FilmArray Meningitis/Encephalitis Panel from BioFire Diagnostics and Meningitis/Encephalitis Panel by PCR from ARUP Laboratories).[6–10] While such assays can rapidly identify H. influenzae (Hi) and Neisseria meningitidis (Nm) species, most do not determine serotype or serogroup. Therefore, it is important for laboratories using assays that do not determine serotype/serogroup to perform either a simultaneous culture or a reflex culture if Hi or Nm is identified. At a minimum, laboratories should collect and maintain an adequate clinical sample for further testing at a laboratory with a PCR assay that can detect serotype/serogroup.
Serotype testing (serotyping)
Serotyping distinguishes encapsulated strains, including Hib, from unencapsulated strains, which cannot be serotyped. The 6 encapsulated types (designated a–f) have distinct capsular polysaccharides that can be differentiated by slide agglutination with type-specific antisera.
To make public health decisions about chemoprophylaxis, microbiology laboratories should perform serotype testing of H. influenzae isolates and clinical specimens that are positive for H. influenzae in a timely manner.[11,12] Even though Hib disease has declined, laboratories should continue routine serotyping. If serotyping is not available at a laboratory, laboratory personnel should contact the state health department. State health departments with questions about serotyping should contact the CDC Meningitis and Vaccine-Preventable Disease Branch laboratory at 404-639-3158.
Molecular typing
Although not widely available, whole genome sequencing (WGS) has been used to type the H. influenzae isolates to assess their genetic similarity. Subtyping the Hib bacterium by pulsed-field gel electrophoresis (PFGE) [13-15] or multilocus sequence typing (MLST) can also be performed for epidemiologic purposes in settings where WGS is not available. Some subtyping methods, such as outer membrane proteins, lipopolysaccharides, or enzyme electrophoresis, are no longer recommended or performed because they were unreliable or too labor intensive. The state health department may direct questions about subtyping to the CDC Meningitis and Vaccine-Preventable Disease Branch laboratory at 404-639-3158.
C. Hepatitis A (see Chapter 3)
Diagnostic tests used to confirm hepatitis A virus (HAV) infection include serologic testing, and occasionally, PCR-based assays to amplify and sequence viral genomes.
Serologic testing
Serologic testing is required to distinguish hepatitis A from other types of viral hepatitis, since clinical or epidemiologic features overlap. Virtually all patients with hepatitis A have detectable immunoglobulin M (IgM) anti-HAV. Acute HAV infection is confirmed during the acute or early convalescent phase of infection by the presence of IgM anti-HAV in serum. Serum for IgM anti-HAV testing should be obtained as soon as possible after onset of symptoms because IgM anti-HAV generally disappears within 6 months after onset of symptoms.
Immunoglobulin G (IgG) anti-HAV appears in the acute or convalescent phase of infection, remains for the lifetime of the person, and confers enduring protection against disease. The antibody test for total anti-HAV measures both IgG anti-HAV and IgM anti-HAV. Persons who are total anti-HAV positive and IgM anti-HAV negative are considered immune, whether from past infection or vaccination history. Tests for IgG anti-HAV are also available.
CDC laboratory special studies
Occasionally, molecular virologic methods such as PCR-based assays are used to amplify and sequence viral genomes. Molecular epidemiologic methods have been useful in understanding HAV transmission within networks of persons with similar risk factors. When applied in combination with conventional epidemiologic methods, HAV sequencing has also been useful in the investigation of outbreaks and determining transmission links[16]. However, for routine surveillance purposes, detection of serologic markers—total anti-HAV, or IgG anti-HAV, and IgM anti-HAV—is sufficient actionable information.
Specimens collected as part of enhanced hepatitis A surveillance in the United States from 2007 through 2013 were sequenced; 472 (62.7%) of 753 available specimens were HAV RNA positive by polymerase chain reaction. Additional specimens from the Food Safety Project A13FBM from 2013–2016 were sequenced: 98 (26.3%) of 373 available specimens were HAV RNA positive. HAV genotypes among these 570 case specimens were: IA (83.2%), IB (16.0%), and IIIA (0.9%) (CDC Division of Viral Hepatitis Laboratory, unpublished data).
Providers with questions about molecular virologic methods should consult with their state health department or the CDC Division of Viral Hepatitis, Laboratory Branch (CDC internal access only.)
D. Hepatitis B (see Chapter 4)
Diagnostic testing for confirmation of hepatitis B virus (HBV) infection includes various serologic markers that can distinguish between different stages of infection and molecular testing, which can provide titer, genotype, and escape mutant information in HBV infected individuals.
Serologic marker testing
Laboratory tests available for evaluation of HBV infection include testing for hepatitis B surface antigen (HBsAg), total antibodies to HBsAg (total anti-HBs); total and IgM antibodies to HBV core antigen (total and IgM anti-HBc) and , and total antibodies to hepatitis B e antigen (total anti-HBe). All these tests are commercially available on automated platforms for use in clinical laboratories. HBV laboratory testing algorithm is complex, and it is frequently done in stepwise fashion, starting with the HBsAg test.
The presence of HBsAg is indicative of ongoing HBV infection and potential infectiousness. In newly infected persons, HBsAg is present in serum 30–-60 days after exposure to HBV. Anti-HBc develops in all HBV infections, appearing at onset of symptoms or liver test abnormalities in acute HBV infection, rising rapidly to high levels, and persisting for life. Acute or recently acquired infection can be distinguished by presence of the IgM class of anti-HBc, which persists for approximately 6 months.
In persons who recover from HBV infection, HBsAg is eliminated from the blood, usually in 2–3 months, and antibodies to hepatitis B surface antigen (anti-HBs) develop. The presence of anti-HBs antibodies at levels equal to or higher than 10 mIU/ml indicates immunity from HBV infection. Anti-HBs antibodies also develop in persons vaccinated against hepatitis B virus. The difference between immunized and previously infected individuals can be determined based on total antibodies to hepatitis B core (total anti-HBc). After recovery from natural infection, most persons will be positive for both anti-HBs and anti-HBc antibodies, whereas only anti-HBs antibodies develop in persons who are successfully vaccinated against hepatitis B. Persons who do not recover from HBV infection and become chronically infected remain positive for HBsAg (and anti-HBc), although a small proportion (0.3% per year) of these persons may eventually clear HBsAg and develop anti-HBs antibodies.[17]
In some cases, anti-HBc is the only serologic marker detected. Isolated anti-HBc can occur after HBV infection in persons who have recovered but whose anti-HBs levels have waned or in persons in whom anti-HBs failed to develop. Certain chronically infected persons may be positive for anti-HBc alone, with HBsAg levels that are below levels detectable by commercially available tests or undetectable by these tests due to S gene mutations. Infants who are born to HBsAg-positive mothers and who do not become infected may also have detectable anti-HBc for up to 24 months after birth from passively transferred maternal antibody.[18]
The diagnosis of acute hepatitis due to hepatitis B virus infection is serologically confirmed by a positive test for IgM antibody to hepatitis B core antigen (IgM anti-HBc). Confirmation of HBsAg-positive results by HBsAg neutralization assay should be done as needed. In addition to acute HBV infection, both perinatal HBV infection and chronic HBV infection are reportable vaccine-preventable conditions. Chronic infection with HBV is confirmed by a positive test for HBsAg accompanied by a negative test for IgM anti-HBc or by 2 positive HBsAg test results that are at least 6 months apart. A diagnosis of perinatal HBV infection is confirmed by a positive test for HBsAg in an infant 1–24 months of age born in the United States or in U.S. territories to an HBsAg-positive mother. Interpretation of hepatitis B serologic test results is summarized in Table 4.
Table 4. Interpretation of hepatitis B serologic tests.
| Serologic Markers | Interpretation | |||
|---|---|---|---|---|
| HBsAg* | Total Anti-HBc† | IgM Anti-HBc§ | Anti-HBs¶ | |
| – | – | – | – | Susceptible, never infected, nor immunized |
| + | – | – | – | Acute infection, early incubation** |
| + | + | + | – | Acute infection |
| – | + | + | – | Acute resolving infection |
| – | + | – | + | Past infection, recovered and immune |
| + | + | – | – | Chronic infection |
| – | + | – | – | False positive (i.e., susceptible), past infection, or “low level” chronic infection |
| – | – | – | + | Immune if titer is ≥10 mIU/ml |
† Antibody to hepatitis B core antigen.
§ Immunoglobulin M.
¶ Antibody to hepatitis B surface antigen.
** Transient HBsAg positivity (lasting less than 18 days) might be detected in some patients during vaccination.
Hepatitis B e antigen (HBeAg) positivity in the serum indicates active viral replication.[19] HBeAg is a secreted product of the pre-core open reading frame (ORF) and is found in both acutely and chronically infected patients. Spontaneous conversion from HBeAg to antibodies to hepatitis B e antigen (anti-HBe) may indicate lower levels of HBV. HBeAg positive mothers are at an increased risk of transmitting HBV infection to their unborn babies, requiring timely preventative intervention soon after birth.[20]
Molecular analysis
Polymerase chain reaction (PCR) based methods are used for determination of HBV viral load (quantitative tests) and HBV DNA presence (qualitative tests): they are available as both commercial Food and Drug Administration (FDA) approved tests as well as laboratory developed tests (LDTs). HBV DNA testing is most commonly used for evaluating a patient with diagnosed HBV infection who is receiving or being considered for treatment; these tests are not typically used for the initial diagnosis of infection. Genotype determination is also possible through PCR-based technology and is available as LDTs, most commonly based on the S gene sequence.[21]
PCR-based methods for amplification or deep sequencing of specific regions of the HBV genome, done in conjunction with epidemiologic studies, may be used for investigating common-source outbreaks of hepatitis B infection, as has been shown for HCV.[22] Furthermore, HBV quasispecies diversity can be useful in evaluation of disease progression. The pre-S region of the HBV genome has been associated with disease progression to hepatocellular carcinoma.[23] In addition, molecular assays are essential for detecting the emergence of vaccine-escape mutants, diagnostic failure mutants, and treatment resistance mutants.[24,25] These types of mutants occur at various rates in different HBV genotypes, so knowing genotype information is essential for diagnosis and treatment of patients. Detection of HBV vaccine escape mutants among vaccinated infants of HBsAg-positive women is important to determine their potential role in vaccine failures.[26] Healthcare professionals with questions about molecular virologic methods or those who identify HBsAg-positive events among vaccinated persons should consult with their state health department or the Epidemiology Branch, Division of Viral Hepatitis, CDC.
E. Influenza (see Chapter 6)
Methods available for the diagnosis of influenza include virus isolation (standard methods and rapid culture assays), molecular detection (reverse transcriptase-polymerase chain reaction [RT-PCR]), detection of viral antigens (enzyme immunoassays [EIA], immunofluorescent antibody [IFA], and commercially available rapid diagnostic kits), and less frequently, use of immunohistochemistry [IHC], electron microscopy, and serologic testing using hemagglutination inhibition or microneutralization.[27,28]
Virus isolation and rapid culture assay
Virus isolation is the gold standard for influenza diagnosis. The following guidelines should be considered.
- Appropriate samples include nasal washes, nasopharyngeal aspirates, nasal and throat swabs, transtracheal aspirates, and bronchoalveolar lavage.
- Samples should be taken within 72 hours of onset of illness to maximize the probability of isolating virus.
- Rapid culture assays that use immunologic methods to detect viral antigens in cell culture are available. These assays can provide results in 18–40 hours, compared with an average of 4–5 days to obtain positive results from standard culture.
Molecular testing methods
RT-PCR, including real-time RT-PCR, can be used to detect the presence of influenza virus in a clinical specimen or to characterize an influenza virus grown in tissue culture or embryonated eggs. This testing can be performed under biosafety level 2 conditions, even for viruses such as avian influenza A(H5N1), which require biosafety level 3 with enhancements for viral culture.
Antigen detection assays
Several methods exist for the diagnosis of influenza infection directly from clinical material:
- Cells from the clinical sample can be stained using an immunofluorescent antibody to look for the presence of viral antigen. Nasal washes, nasopharyngeal aspirates, nasal and throat swabs, gargling fluid, transtracheal aspirates, and bronchoalveolar lavage are suitable clinical specimens.
- Commercially available kits to test for the presence of viral antigens fall into 3 groups: the first detects only influenza type A viruses; while the second detects both influenza type A and B viruses but does not differentiate between virus types; and the third detects both influenza type A and B viruses and distinguishes between the two. Results of these rapid antigen detection tests can be available in less than 1 hour, although these tests are usually less sensitive (generally approximately 50%–70%) than RT-PCR testing.[29-32]
- Other less frequently used methods include immunostaining and visualization of viral antigens by electron microscopy. This method and RT-PCR methods may also be used for detection of influenza antigens and nucleic acids, respectively, in postmortem respiratory tissue samples.
When direct antigen detection methods are used for the diagnosis of influenza, it is important to collect and reserve an aliquot of the clinical sample for possible further testing. The medium used to store the specimen for some rapid testing methods is inappropriate for viral culture; in this case, it is necessary to collect 2 separate samples. These additional or reserved samples may be used to confirm direct test results by culture and to subtype influenza A isolates.
Serologic testing
Serologic diagnosis of influenza infection requires paired serum specimens. The acute-phase sample should be collected within 1 week of the onset of illness, and the convalescent-phase sample should be collected approximately 2–3 weeks later.
Hemagglutination inhibition (HI) tests are the preferred method of serodiagnosis. A positive result is a 4-fold or greater rise in titer between the acute- and convalescent-phase samples when tested at the same time. Serologic test results are usually available in 24 hours.
Serologic testing is most useful in special studies; serologic diagnosis of influenza is not used for national surveillance because of the lack of standardized testing methods and interpretation.
F. Measles (see Chapter 7)
Serologic testing
Serologic testing for antibodies to measles is widely available. Generally, in a susceptible person exposed to wild-type measles virus, the IgM response begins around the time of rash onset and can be detected for 1-2 months. The IgG response starts more slowly, at about 5-10 days after rash onset, but typically persists for a lifetime. The diagnosis of acute measles infection can be made by detecting IgM antibody to measles in a single serum specimen or by detecting seroconversion — a 4-fold rise in the titer of IgG antibody in 2 serum specimens obtained approximately 2 weeks apart. Uninfected persons are IgM negative, but will either be IgG negative or IgG positive, depending upon their previous disease or vaccination histories.
Recommendations for serologic testing for measles
- An EIA test for IgM antibody to measles in a single serum specimen, obtained at the first contact with the suspected measles case-patient, is one of the recommended methods for diagnosing acute measles.
- A single-specimen test for IgG is the most commonly used test for immunity to measles because IgG antibody is long-lasting.
- Testing for IgG along with IgM is recommended for suspected measles cases.
- Paired sera (acute and convalescent) may be tested for seroconversion or a 4-fold rise in IgG antibody to measles to confirm acute measles infection.
- When a patient with suspected measles has been recently vaccinated (6-45 days prior to rash onset), neither IgM nor IgG antibody responses can distinguish measles disease from rash following vaccination. In this instance, a viral throat or nasopharyngeal swab specimen should be obtained so CDC can attempt to distinguish between vaccine virus and wild-type virus (Table 5).
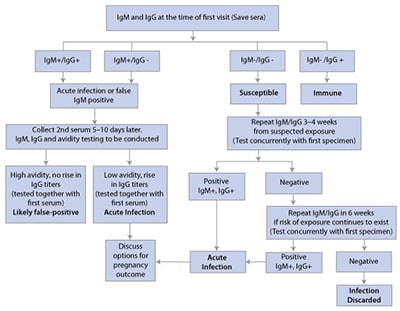
Table 5. Interpretation of measles enzyme immunoassay results*
| IgM Result | IgG Result | Previous infection history | Current infection | Comments |
|---|---|---|---|---|
| + | – or + | Not vaccinated, no prior history of measles | Recently received first dose of measles vaccine | Seroconversion. IgG response depends on timing of specimen collection. |
| + | – or + | Not vaccinated, no prior history of measles | Wild-type measles | Seroconversion. Classic clinical measles. IgG response depends on timing of specimen collection. |
| + | – or + | Previously vaccinated, primary vaccine failure | Recently received second dose of measles vaccine | Seroconversion. IgG response depends on timing of specimen collection. |
| – | + | Previously vaccinated, IgG+ | Recently received second dose of measles vaccine | IgG level may stay the same or may boost. |
| + | + | Previously vaccinated, IgG+ | Wild-type measles | May have few or no symptoms (e.g., no fever or rash). |
| + | + | Recently vaccinated | Exposed to wild-type measles | Cannot distinguish between vaccine or wild-type virus; evaluate on epidemiologic grounds.† |
| – | + | Distant history of natural measles | Vaccine | IgG level may stay the same or may boost. |
| + (at least in some patients) |
+ | Distant history of natural measles | Wild-type measles | May have few or no symptoms. |
† However, in this circumstance, IgM testing will be helpful. If negative, it could rule out wild-type measles infection.
Tests for IgM antibody. Although multiple possible methods exist for testing for IgM antibody, EIA is the most consistently accurate test and is therefore the recommended method. There are 2 formats for IgM tests. The first and most widely available is the indirect format, which requires a specific step to remove IgG antibodies. Problems with removal of IgG antibodies can lead to false-positive[33] or, less commonly, false-negative results.
The second format, IgM capture, does not require the removal of IgG antibodies. This is the preferred reference test for measles.
EIA tests for measles are often positive on the day of rash onset. However, in the first 72 hours after rash onset, up to 30% of tests for IgM may give false-negative results. Tests that are negative in the first 72 hours after rash onset should be repeated; serum should be obtained for repeat testing 72 hours after rash onset. IgM is detectable for at least 28 days after rash onset and frequently longer.[34]
When a laboratory IgM test result is suspected of being false-positive, additional tests may be performed. False-positive IgM results for measles may be due to the presence of rheumatoid factor in serum specimens. Serum specimens from patients with other rash illness, such as parvovirus B19, rubella, and roseola, have been observed to yield false-positive reactions in some IgM tests for measles. False-positive tests may be suspected when thorough surveillance reveals no source or spread of cases, when the case does not meet the clinical case definition, or when the IgG result is positive within 3 days of rash onset. In these situations, confirmatory tests may be done at the state public health laboratory or at CDC. IgM results by tests other than EIA can be validated with EIA. Indirect EIA tests may be validated with capture EIA.
Tests for IgG antibody. Because tests for IgG require 2 serum specimens and a confirmed diagnosis cannot be made until the second specimen is obtained, IgM tests are generally preferred. However, if the IgM tests remain inconclusive, a second (convalescent-phase) serum specimen, collected 14-30 days after the first (acute-phase) specimen, can be used to test for an increase in the IgG titer. These tests can be performed in the state laboratory or at CDC. A variety of tests for IgG antibodies to measles are available; these include EIA, indirect fluorescent antibody tests, and plaque reduction neutralization. Complement fixation, although widely used in the past, is no longer recommended. The “gold standard” test for serologic evidence of recent measles virus infection is plaque reduction neutralization test of IgG in acute- and convalescent-phase paired sera.
Paired IgG testing for laboratory confirmation of measles requires the demonstration of a 4-fold rise in titer of antibody against measles. The tests for IgG antibody should be conducted on both acute- and convalescent-phase specimens at the same time. The same type of test should be used on both specimens. The specific criteria for documenting an increase in titer depend on the test. EIA values are not titers and increases in EIA values do not directly correspond to rises in titer.
RT-PCR and virus isolation
Isolation of measles virus in culture or detection of measles virus by RT-PCR in clinical specimens confirms the diagnosis of measles. Since virus isolation can take weeks to perform, RT-PCR is preferred for case confirmation. It is important to note that a negative culture or RT-PCR result does not rule out measles because test results are greatly affected by the timing, quality, and handling of the clinical specimens. If measles virus is isolated or detected by RT-PCR, the viral genotype can be used for molecular epidemiology and to distinguish between measles disease caused by a wild-type measles virus and a measles vaccine reaction.
Viral isolation and RT-PCR are important for molecular epidemiologic surveillance to help determine:
- the origin of the virus,
- which viral strains are circulating in the United States, and
- whether these viral strains have become endemic in the United States.
Specimens (urine, nasopharyngeal aspirates, heparinized blood, or throat swabs) from clinically suspected cases of measles obtained for virus isolation should be shipped to the state public health laboratory or to CDC at the direction of the state health department as soon as measles is confirmed. Specimens should be properly stored while awaiting case confirmation (see Appendix 7 [2 pages]). Clinical specimens for virus isolation should be collected at the same time as samples for serologic testing. Because virus is more likely to be isolated when the specimens are collected within 3 days of rash onset, collection of specimens for virus isolation should not be delayed until laboratory confirmation is obtained. Clinical specimens should ideally be obtained within 7 days of rash onset and should not be collected if more than 10 days have passed after rash onset.
G. Meningococcal disease, Neisseria meningitiditis (see Chapter 8)
Identification of N. meningitidis
Presumptive identification by Gram stain: The Gram stain is an empirical method for differentiating bacterial species into 2 large groups based on the chemical and physical properties of their cell walls. Gram-positive bacteria retain the primary stain while gram-negative bacteria take the color of the counterstain. A Gram stain can also serve to assess the quality of a clinical specimen. Gram staining for N. meningitidis is commonly used and continues to be a reliable and rapid method for presumptive identification. Intracellular gram-negative diplococci in CSF can be considered meningococci until proven otherwise.
Presumptive identification by antigen detection: Latex agglutination can be used for rapid detection of meningococcal capsular polysaccharides in CSF; however, false-negative or false-positive results can occur. Antigen agglutination tests on serum or urine samples are unreliable for the diagnosis of meningococcal disease.[35]. Immunohistochemistry can be used for meningococcal detection in formalin-fixed tissues.
Confirmation by culture and PCR: The case definition for confirmed meningococcal disease requires isolation of N. meningitidis or detection of Nm. DNA from a normally sterile site. Typically, normally sterile sites include blood or CSF, but can also include joint, pleural, or pericardial fluid. Aspirates or skin biopsies of purpura or petechiae can yield meningococci in cases of meningococcemia. The typical media used to grow the organism are chocolate agar or Mueller-Hinton medium in an atmosphere containing 5% carbon dioxide.[36]
Real-time PCR detects DNA of meningococci in blood, CSF, or other clinical specimens and is an acceptable alternative method for confirming meningococcal cases. Real-time PCR assays are available to detect DNA of N. meningitidis and all 6 serogroups in blood, CSF, or other clinical specimens.[6-10] A major advantage of PCR is that it allows for detection of N. meningitidis from clinical samples in which the organism could not be detected by culture methods, such as when a patient has been treated with antibiotics before a clinical specimen is obtained for culture. Even when the organisms are nonviable following antimicrobial treatment, PCR can still detect N. meningitidis DNA.[37] Because of the severity of meningococcal disease, it is critical to treat the patient as soon as infection is suspected and not delay to obtain a culture or laboratory results.
Several commercial multiplex PCR assays capable of simultaneously testing a single specimen for an array of pathogens that cause blood infections or meningitis/encephalitis are now available, primarily for clinical settings (e.g., FilmArray Blood Culture Identification Panel and FilmArray Meningitis/Encephalitis Panel from BioFire Diagnostics and Meningitis/Encephalitis Panel by PCR from ARUP Laboratories).[9,38,39] While such assays can rapidly identify Hi and Nm species, most do not determine serotype or serogroup. Therefore, it is important for laboratories using assays that do not determine serotype/serogroup to perform either a simultaneous culture or a reflex culture if Hi or Nm is identified. At a minimum, laboratories should collect and maintain an adequate clinical sample for further testing at a laboratory with a PCR assay that can detect serotype/serogroup results.
Serogroup testing
Serogrouping distinguishes encapsulated meningococcal strains from unencapsulated strains, which do not express any capsule on cell surface and cannot be serogrouped. The 12 encapsulated groups (A, B, C, W, X, Y, etc.) have distinct capsular polysaccharides that can be differentiated by slide agglutination with specific antisera or PCR.
Microbiology laboratories should perform serogroup testing of meningococcal isolates and clinical specimens that are positive for N. meningitidis in a timely manner. If serogrouping is not available at a laboratory, laboratory personnel should contact the state health department. State health departments with questions about serogrouping should contact the CDC Meningitis and Vaccine Preventable Disease Branch laboratory at 404-639-3158.
Susceptibility testing
Routine antimicrobial susceptibility testing of meningococcal isolates is not recommended. N. meningitidis strains with decreased susceptibility to penicillin G have been identified sporadically from several regions of the United States, Europe, and Africa.[40] Most of these isolates with reduced penicillin susceptibility remain moderately susceptible (minimum penicillin inhibitory concentration of between 0.12 µg/mL and 1.0 µg/mL). High-dose penicillin G remains an effective treatment against moderately susceptible meningococci. Surveillance of susceptibility patterns in populations should be conducted to monitor trends in N. meningitidis susceptibility.
Molecular typing during outbreaks
Phenotypic and genotypic methods are used to investigate meningococcal diversity. Capsular polysaccharide (serogroup), porin protein PorB (serotype), and porin protein PorA (serosubtype) are all phenotypic characteristics that can be used to distinguish meningococci from one another.[41] Outbreaks of meningococcal disease are usually caused by the same or closely related strains.[42] Molecular genotyping techniques such as PFGE, 16S rRNA gene sequencing, or multilocus sequence typing (MLST) are used for subtype characterization of an outbreak clone.[43,44] This subtyping helps to better define the extent of the outbreak. It is crucial to have rapid and reliable results in determining the meningococcal serogroup in an outbreak because public health response will differ for vaccine-preventable or non-vaccine–preventable disease. Molecular genotyping provides important tools for understanding the overall epidemiology of meningococcal disease, but different methods may be more useful in certain settings. PFGE or 16S rRNA gene typing seem to be most useful for outbreak and short-time-period epidemiology, whereas MLST has become the “gold standard” for long-term, global tracing of meningococcal population changes.
H. Mumps (see Chapter 9)
Fully vaccinated persons can become infected with mumps but are at much lower risk for mumps illness and mumps complications. Recent mumps outbreaks have most commonly occurred among fully vaccinated populations, with illness more common in persons who have prolonged, close contact exposures, such as sharing water bottles or cups, kissing, practicing sports together or living in close quarters with a person who has mumps. Laboratory confirmation of mumps can be more challenging among vaccinated persons because the viral load may be lower and less easily detected [45] and the IgM response may be absent, delayed, or short-lived compared with unvaccinated people. [46,47]
Successful detection of mumps virus by real-time reverse transcription polymerase chain reaction (rRT-PCR) or a mumps IgM response is dependent upon proper collection (i.e., parotid massage for buccal* swabs), timing of collection, and proper transport and storage of specimens. Failure to detect mumps virus RNA by rRT-PCR or to detect IgM in serum in samples from a person with clinically compatible mumps does not rule out mumps as a diagnosis. Efforts to train providers in proper collection, storage and shipment of specimens have improved mumps detection. [46,48]
* The buccal mucosa is the lining of the cheeks
Acute mumps infection can be confirmed by the detection of mumps virus RNA, by rRT-PCR, or by isolation of mumps virus from a clinical specimen. A case with a positive mumps rRT-PCR result or virus isolation is classified as confirmed. Maximal viral shedding occurs 1–3 days prior to onset and through day 5 following onset of parotitis. Early collection (0 to 3 days since parotitis onset) of a buccal swab provides the best means of laboratory confirmation, particularly among persons with a history of vaccination since virus titers may be lower and virus may be cleared more rapidly than in unvaccinated persons. [45,47,49]
If mumps RNA is detected in buccal swab samples, the samples can be submitted for sequence analysis to determine the viral genotype. Analysis of sequence data can help to monitor viral transmission. Additionally, in cases of suspected vaccine reactions (i.e., patients with suspected mumps who were recently vaccinated), it is necessary to collect buccal swab samples for virus detection and genotyping. Since serologic tests cannot differentiate between an exposure to vaccine and an exposure to wild-type mumps virus, genotyping must be used to confirm detection of vaccine strain or wild-type virus.
Buccal swab specimens should be obtained by massaging the parotid gland area for 30 seconds prior to swabbing the area around Stensen’s duct. A commercial product designed for the collection of throat specimens or a flocked polyester fiber swab can be used. Synthetic swabs are preferred over cotton swabs, which may contain substances that are inhibitory to enzymes used in rRT-PCR. Flocked synthetic swabs appear to be more absorbent and elute samples more efficiently. See the CDC mumps laboratory website for information on viral transport medium, specimen storage, and shipment recommendations.
The presence of serum mumps IgM can also be used to aid in the diagnosis of mumps infection but is not confirmatory. Detection of mumps IgM is improved if serum specimens are collected greater than 3 days after parotitis onset.[46,50,51] A clinically compatible case with a positive serum anti-mumps immunoglobulin M (IgM) antibody result is classified as probable.
Specimen Collection
Appropriate timing and collection of specimens is important to improve the sensitivity of laboratory testing. Mumps testing job-aids for epidemiologically linked and sporadic cases are here.
- The parotid should be massaged for 30 seconds prior to buccal swab collection. For instructions on how to appropriately collect a buccal swab for mumps testing, a guide is provided here.
- If it has been ≤3 days since parotitis onset, only collect a buccal swab specimen for rRT-PCR.
- If it has been >3 days since parotitis onset, collect a buccal swab specimen for rRT-PCR and a serum specimen for IgM detection.
- If the patient has orchitis/oophoritis, mastitis, pancreatitis, hearing loss, meningitis or encephalitis without known cause and does not have parotitis, collect a buccal swab specimen for rRT-PCR, a urine specimen* for rRT-PCR, and a serum specimen for IgM detection regardless of the days since symptom onset.
*CDC currently only accepts buccal swabs, check with your local laboratory/VPD reference center to see what sample types are acceptable.
For individuals presenting with symptoms of mumps without known epidemiologic-linkage, multiplex testing for other infectious etiologies is recommended concurrent with mumps testing, including:
- Influenza virus (influenza A virus subtype H3N2)
- Parainfluenza virus types 1–3
- Epstein-Barr virus
- Human herpesviruses 6A and 6B
- Herpes simplex viruses 1 and 2
- Coxsackievirus A
- Adenovirus
- Parvovirus B19
- Echovirus
- Lymphocytic choriomeningitis virus
- HIV
- Bacteria including Staphylococcus aureus and alpha hemolytic streptococcus
Virus Isolation and rRT-PCR
To provide state and local jurisdictions with enhanced surveillance testing capacity for 7 vaccine-preventable diseases (VPDs) ― mumps, measles, rubella, varicella zoster virus, Bordetella pertussis, Haemophilus influenzae, and Neisseria meningitidis — CDC, in collaboration with the Association of Public Health Laboratories (APHL) and the Council of State and Territorial Epidemiologists (CSTE), in 2013 established 4 Vaccine Preventable Disease Reference Centers (VPD-RCs). These VPDs are typically detected by laboratory testing such as rRT-PCR assay of clinical specimens. The VPD-RCs provide state and local health departments with surge capacity for large outbreaks, testing for jurisdictions without rRT-PCR capabilities, and genotyping. Since their establishment, VPD-RCs have received and tested the majority of the mumps clinical specimens.
Mumps is laboratory confirmed by rRT-PCR or viral culture from buccal or urine specimens. Collect buccal swab samples as soon as mumps disease is suspected. rRT-PCR has the greatest diagnostic sensitivity when samples are collected ≤3 days of parotitis onset. Urine is not the optimal specimen for rRT-PCR and is not preferred as a specimen for patients with parotitis, even for those patients presenting more than 3 days since symptom onset.[48,52-57] Testing urine during an outbreak does not significantly increase diagnostic yield and can negatively impact testing resources. However, in patients presenting with mumps complications (orchitis, oophoritis, nephritis, encephalitis, meningitis) without parotitis or other salivary gland swelling collecting a urine specimen in addition to a buccal specimen might help to confirm mumps infection. CDC is currently only accepting buccal swabs, check with your local laboratory/VPD reference center to see what sample types are acceptable.
At the onset of a suspected mumps outbreak, patients suspected to have mumps should be tested by rRT-PCR to confirm mumps and rule out other possible etiologies. However, once a mumps outbreak is confirmed, jurisdictions can consider alternate strategies to reduce testing to conserve public health resources (See Strategies for the Control and Investigation of Mumps Outbreaks).
Molecular Typing
Molecular epidemiologic surveillance makes it possible to build a sequence database that will help monitor any changes in the genotype prevalence over time and help track transmission of mumps strains circulating in the United States. Collecting and submitting specimens for sequencing is important for molecular surveillance and informing public health. Specimens for molecular typing should be obtained as soon as possible after the onset of parotitis, ideally from the day of onset to 3 days later (not more than 10 days after parotitis onset).
Molecular typing is the only test that can discriminate vaccine reactions from wild-type infections. Specific instructions for specimen collection and shipping are available from the CDC mumps laboratory website.
CDC and the VPD-RCs can perform mumps genotyping on rRT-PCR–positive specimens. The nucleotide sequence of the entire mumps short hydrophobic (SH) gene is used to assign mumps viruses to one of 12 recognized genotypes.[58] In some circumstances, a genotype has been associated with endemic circulation of mumps virus in a country; however, routine genotype surveillance for mumps is limited to only a few countries. The genetic information from circulating mumps viruses is used to track the transmission pathways of the virus and can be used to suggest epidemiologic links, or lack thereof, between cases and outbreaks.[59]
CDC has detected mostly genotype G among people with mumps in the United States since we initiated routine genotype surveillance for mumps in 2006.[60] Mumps outbreaks are typically associated with only one genotype. There are no differences in the genotypes detected in vaccinated and unvaccinated people who have gotten mumps in the United States.
Public health laboratories are encouraged to send patient samples from rRT-PCR positive sporadic cases of mumps, as well as select specimens from an outbreak, (for more information on which specimens, see Strategies for the Control and Investigation of Mumps Outbreaks) to CDC or the VPD-RC’s for genotyping. If the outbreak spreads to another community, an effort should be made to obtain a genotype on initial positive specimens from the new outbreak.
Molecular surveillance of mumps outbreaks
- To monitor ongoing established outbreaks, a maximum of 5 specimens per week from these outbreaks should be sent to a VPD-RC or CDC for sequencing.
- Obtain an SH sequence for cases at the beginning of an outbreak, during each epidemiologic week of the outbreak, and if/when the outbreak spreads to a new area or community.
- In an outbreak, obtaining a sequence on every specimen is not necessary especially when linkages between cases are well understood.
- Obtaining sequences from sporadic cases is important because they help us monitor transmission patterns of mumps virus.
- Identifying travel and/or a potential source for mumps cases will help us understand the global distribution of mumps genotypes.
- Jurisdictions that perform mumps rRT-PCR testing at their state or local public health laboratories should also send rRT-PCR positive specimens to monitor new or ongoing outbreaks.
- Global virologic surveillance for mumps is not as robust as for measles or rubella, so the true diversity of genotypes is not known.
Priority patients for sequencing:
- Patients with parotitis, salivary gland swelling, oophoritis, or orchitis who are not epidemiologically linked to a confirmed or probable mumps case-patient (sporadic cases)
- Patients with aseptic meningitis, encephalitis, hearing loss, mastitis, or pancreatitis in the absence of parotitis, salivary gland swelling, oophoritis, or orchitis (severe mumps complications)
- Patients who traveled internationally during their likely incubation period (12–25 days prior to onset)
- Patients with recurrent parotitis (submit samples from both occurrences when possible)
- Patient who received ≥3 doses of MMR more than 28 days before symptom onset
Serologic Testing for IgM Antibody
Vaccination status and timing of specimen collection affect the ability to detect IgM in persons infected with mumps. In general, the percentage of specimens with a positive IgM result is highest in unvaccinated persons, intermediate in one-dose vaccine recipients, and lowest in 2-dose vaccine recipients. Specimens collected greater than 3 days post-parotitis onset are more likely to have a positive IgM result.
IgM test methods and kits vary considerably in sensitivity and specificity, with some indirect EIA and immunofluorescent assays detecting as few as 12%–15% of confirmed mumps cases.[46] IgM capture ELISA is the most sensitive serologic method detecting 46%–71% of rRT- PCR confirmed cases.[46] The local or state health department can provide guidance on available laboratory services and preferred tests.
Failure to detect mumps IgM in previously vaccinated persons who are infected with mumps has been well documented. People with a history of mumps vaccination may not have detectable mumps IgM antibody regardless of timing of specimen collection, and there are also concerns with cross-reactivity to other viruses.
Enzyme immunoassay. EIA is a highly specific test for diagnosing acute mumps infection. Commercial immunofluorescence (IFA) and indirect EIA tests are less sensitive than capture IgM assays.[45] At the direction of the state health department, healthcare providers and state and local health departments may send serum specimens from persons with suspected mumps cases to the CDC Viral Vaccine-Preventable Diseases Branch for IgM detection by their in-house Capture EIA.
Immunofluorescence assays. IFAs have the advantage of being relatively inexpensive and simple. The reading of IFA IgM tests requires considerable skill and experience since nonspecific staining may cause false-positive readings. IFA and indirect EIA may be susceptible to interference by high levels of IgG. Treatment of serum to remove IgG may be necessary to avoid false-positive results.
Note: Commercially available IFA antibody assays and EIA kits for detection of mumps IgM are currently not approved by the U.S. Food and Drug Administration. Each laboratory must validate these tests independently.
Serum collection and timing of the mumps IgM response
Unvaccinated persons: IgM antibody is detectable within 5 days after onset of symptoms, reaches a maximum level about a week after onset, and remains elevated for several weeks or months.[61,62] Most IgM detection assays work well in unvaccinated persons although false negative results may occur if collected too early.[46] If an acute-phase serum sample collected ≤3 days after parotitis onset is negative for IgM and negative by rRT-PCR, testing a second serum sample collected on day 5 or later after symptom onset is recommended as the IgM response may require more time to reach a detectable level.
Vaccinated persons: Patients that mount a secondary immune response to mumps, as seen in most previously vaccinated persons, may not have an IgM response or it may be absent, delayed or short-lived depending on the timing of specimen collection.[61] Because of this, a high number of false-negative results may occur in previously vaccinated individuals if specimens are collected ≤ 3 days. False-positive IgM results can also occur and appear to be more prevalent with certain IgM test formats, such as the IFA. Collecting specimens >3 days after parotitis onset improves the ability to detect IgM. During outbreaks, 13%–46% of serum samples collected from patients less than 3 days after symptom onset were positive compared with 71% of serum samples collected >3 days.[46] However, persons with a history of mumps vaccination may not have detectable mumps IgM antibody regardless of the timing of specimen collection and failure to detect mumps IgM in serum in samples from a person with clinically compatible mumps does not rule out mumps as a diagnosis.
Serologic Testing for IgG Antibody
A variety of tests for IgG antibodies to mumps are available and include EIA, IFA, and plaque reduction neutralization. The specific criteria for documenting the presence of mumps IgG antibody or an increase in titer depends on the test.
Adults born in 1957 or later or persons with no history of mumps illness or vaccination may have detectable mumps IgG due to a previous asymptomatic infection.
Diagnosis of mumps with IgG: Documentation of seroconversion from IgG negative to IgG positive by EIA or a 4-fold rise in IgG titer using paired specimens as measured in plaque-reduction neutralization assays or similar quantitative assays can be used to aid in the diagnosis of mumps. Because confirmation of mumps via seroconversion requires two visits at least 2 weeks apart, this method is not practical for immediate case management and confirmation, and faster methods of case confirmation, such as rRT-PCR, are preferred. Tests for IgG antibody should be conducted on both acute- and convalescent-phase specimens at the same time, and the same type of test should be used on both specimens. EIA values are not titers and increases in EIA values do not directly correspond to rises in titer results.
- Unvaccinated persons: In unvaccinated persons, IgG antibody increases rapidly after onset of symptoms and is typically long lasting. IgG may be present in the acute-phase sample depending on the timing of sample collection.
- Vaccinated persons: In vaccinated persons, the IgG will likely already be elevated in the acute-phase blood sample, which frequently prevents detection of a 4-fold rise in IgG titer in the convalescent serum specimen. For this reason, checking for a 4-fold rise in IgG is not recommended for use among previously vaccinated persons.
Presumptive Laboratory Evidence of Immunity
The presence of mumps-specific IgG, as detected using a serologic assay (EIA or IFA), is considered evidence of prior exposure to mumps vaccine or mumps virus but does not necessarily predict the presence of neutralizing antibodies or protection from mumps disease. There is no established correlate of protection for mumps.
I. Pertussis (see Chapter 10)
Culture
Isolation of B. pertussis by bacterial culture remains the gold standard for diagnosing pertussis. It is also required for antimicrobial susceptibility testing and molecular typing. Although bacterial culture is specific for the diagnosis, it is relatively insensitive. Under optimal conditions 80% of suspected cases in outbreak investigations can be confirmed by culture; in most clinical situations isolation rates are much lower.[63] The timing of specimen collection can affect the isolation rate, as can inadequately collected specimens and concurrent use of effective antimicrobial agents. Because patients can remain culture positive even while taking effective antibiotics (e.g., when strains are resistant to the antibiotic), nasopharyngeal swab for culture should be obtained regardless of concurrent use of an antibiotic.
Fastidious growth requirements make B. pertussis difficult to isolate. Isolation of the organism using direct plating is most successful during the catarrhal stage (i.e., first 1–2 weeks of cough). All persons with suspected pertussis disease should have a nasopharyngeal aspirate or swab obtained from the posterior nasopharynx for culture. Recovery rates of B. pertussis from nasopharyngeal aspirates are similar to or higher than rates of recovery from nasopharyngeal swabs;[63-66] rates of recovery from throat and anterior nasal swabs are unacceptably low. Therefore, specimens should be obtained from the posterior nasopharynx, not the throat (Figure 1), by using a polyester (such as Dacron) or nylon flocked nasopharyngeal swab. Cotton-tipped swabs are not acceptable as residues present in these materials inhibit growth of B. pertussis. Specimens should be plated directly onto selective culture medium or placed in transport medium. Regan-Lowe agar or Bordet-Gengou agar are generally used for culture; half-strength Regan-Lowe should be used as the transport medium. Success in isolating the organism declines if the patient has received prior antibiotic therapy effective against susceptible B. pertussis (erythromycin or trimethoprim-sulfamethoxazole), if there is a delay in specimen collection beyond the first 2 weeks of illness, or if the patient has been vaccinated. A positive culture for B. pertussis confirms the diagnosis of pertussis. For this reason, access to a microbiology laboratory that is prepared to perform this service for no cost or for limited cost to the patient is a key component of pertussis surveillance.
Polymerase chain reaction
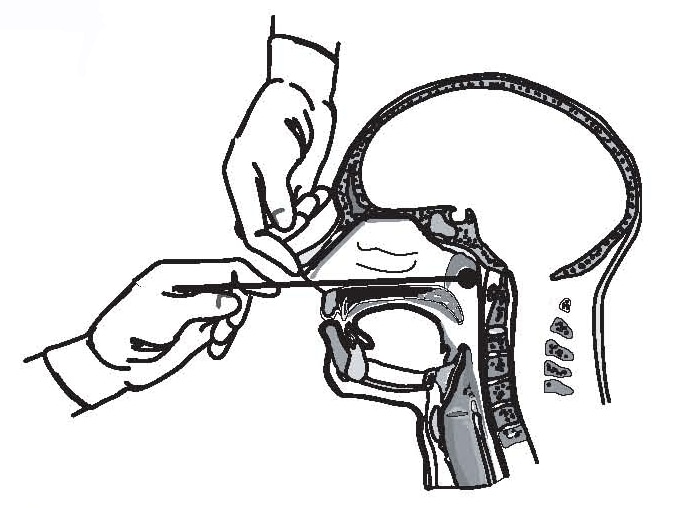
Figure 1: Proper technique for obtaining a nasopharyngeal specimen for isolation of B. pertussis
PCR is an important tool for timely diagnosis of pertussis and is widely available to clinicians. PCR is a molecular technique used to detect DNA sequences of the Bordetella pertussis bacterium, and unlike culture, does not require viable (live) bacteria present in the specimen.[67,68] PCR testing of nasopharyngeal swabs or aspirates can be a rapid, sensitive, and specific method for diagnosing pertussis;[68,69] however, false-positive results may be obtained because of contamination in the laboratory or during specimen collection.[69,70] Although early signs and symptoms of pertussis are often non-specific, only patients with signs and symptoms consistent with pertussis should be tested. Asymptomatic contacts of confirmed cases should not be tested and testing of contacts should not be used for postexposure prophylaxis decisions.
Since its inclusion in the case definition in 1997, the proportion of cases confirmed by PCR has increased substantially, and many laboratories now use only PCR to confirm pertussis [71]. However, the assay varies among laboratories and is not standardized[69] and assay procedures, as well as sensitivity and specificity, can vary greatly between laboratories. Thus, interpretation criteria for diagnosis vary. Interpretation of PCR results, especially those with high cycle threshold (Ct) values should be done in conjunction with an evaluation of signs and symptoms and available epidemiological information. For more information about interpretation of PCR Ct values, see Best Practices for Health Care Professionals on the use of Polymerase Chain Reaction (PCR) for Diagnosing Pertussis. [72]
While PCR is increasingly used as the sole diagnostic test for pertussis, CDC recommends that PCR be used in conjunction with culture when feasible, rather than as an alternative test. Direct comparison with culture is necessary for validation, and the use of multiple targets is recommended to distinguish B. pertussis from other Bordetella species.[73] Even if a laboratory has validated its PCR method, isolation of B. pertussis by culture should be attempted when possible. B. pertussis isolates can then be evaluated for azithromycin and erythromycin susceptibility and by molecular typing methods, which can help define the molecular epidemiology of strains circulating in the United States.
The timing of PCR testing for pertussis can significantly affect its ability to accurately diagnose the disease. PCR has optimal sensitivity during the first 3 weeks of cough when bacterial DNA is still present in the nasopharynx. After the fourth week of cough, the amount of bacterial DNA rapidly diminishes, which increases the risk of obtaining falsely negative results.
Collection methods for PCR are similar to those for culture, and often the same sample can be used for both tests. However, calcium alginate swabs cannot be used to collect nasopharyngeal specimens for PCR. Swabs used solely for PCR (and not culture) testing may be placed in a dry, sterile tube for transport to the laboratory. Use of liquid transport media is discouraged due to the risk of specimen contamination.
Serologic testing
Although serologic testing has proved useful in clinical studies and outbreak investigations, it is not yet standardized. Also, the lack of association between antibody levels and immunity to pertussis makes results of serologic testing difficult to interpret. Even though serology is not part of the clinical case definition, commercial clinics and manufacturers still provide serologic assays and serology kits for pertussis diagnosis. No serologic assay is FDA approved and very little is understood about the clinical accuracy of these commercially available serologic assays.
Cutoff points for diagnostic values of immunoglobulin (Ig)G antibody to pertussis toxin (PT) have not been established, and current IgA and IgM assays lack adequate sensitivity and specificity. In the absence of recent immunization, an elevated serum IgG antibody to PT after 2 weeks of cough onset is suggestive of recent B. pertussis infection. An increasing titer or a single IgG anti-PT value of approximately 100 IU/mL or greater (using standard reference sera as a comparator) can be used for diagnosis.
Only in Massachusetts, where the state utilizes its own clinically validated assay, is serology used for clinical diagnosis and reporting for patients 11 years of age and older.[74] CDC, together with the FDA, validated an IgG anti-PT ELISA that has proven useful for diagnosis in adolescents and adults during the later phases of the disease.[75-77] The assay is currently performed at the Minnesota Department of Health Laboratory to support other state health departments, as part of the Vaccine Preventable Diseases Reference Centers Program.[74]
Elsewhere, with few exceptions, it is not known if serologic testing has been appropriately validated or standardized. Therefore, serologic testing should not be relied upon to confirm cases for national reporting purposes. Cases meeting the clinical case definition that are serologically positive, but not culture positive or PCR positive, should be reported as probable cases.
Direct fluorescent antibody testing
DFA testing of nasopharyngeal secretions, which is sometimes used to screen for pertussis, is not recommended as a test for diagnosing pertussis. A positive DFA result may increase the probability that the patient has pertussis, but it has limited specificity (frequent false-positive results) and is not a confirmatory test. A monoclonal DFA test is available, but the sensitivity and specificity are variable.
Elevated white blood cell count
An elevated white blood cell count with a lymphocytosis (i.e., increase in lymphocyte count) is usually present in cases of pertussis. The absolute lymphocyte count can reach 20,000/mm3 or higher. However, there may be no lymphocytosis in very young infants, vaccinated children, or adults with mild cases of pertussis. The white blood cell count is not a test of confirmation.
Pulsed-field gel electrophoresis
PFGE is a type of DNA fingerprinting. This technique has been a useful tool for distinguishing epidemiologically related strains (e.g., strains from the same household or small community), while showing diversity within larger geographic areas such as cities, counties, and states.[78,79]
Multi-locus variable-number tandem repeat analysis
Multi-locus variable-number tandem repeat analysis (MLVA) is a molecular typing method that compares genomic regions of direct repeats (variable-number tandem repeats, or VNTRs) between strains; (80) it demonstrates less diversity of strains than PFGE. MLVA has the advantage of being applicable to both cultures and directly to nucleic acid extracted from clinical specimens (81).
Multi-locus sequence typing
Multi-locus sequence typing (MLST) is another typing method that analyzes nucleotide variation in a predetermined set of genes or gene fragments (82). MLST methods vary between bacterial species. Although an MLST method for B. pertussis is not standardized, many laboratories analyze fragments of 3 genes and promoter regions encoding virulence factors that are included in several acellular pertussis vaccines.(83,84)
State health departments with questions regarding isolation, PCR detection, serology, or other test methods for B. pertussis should contact the CDC Pertussis and Diphtheria Laboratory at 404-639-1231.
J. Pneumococcal infection (see Chapter 11)
Culture
Streptococcus pneumoniae is a gram-positive, lancet-shaped diplococcus that commonly inhabits the throat as normal flora. S. pneumoniae commonly causes lower and upper respiratory diseases, including pneumonia, meningitis, and acute otitis media. Diagnosis of invasive pneumococcal infection is confirmed by culture and isolation of S. pneumoniae from a normally sterile body site (e.g., blood, CSF, pleural fluid, peritoneal fluid). Alternatively, diagnosis can be confirmed from culture-negative specimens from normally sterile sites using real-time PCR.
Antibiotic resistance
The Clinical Laboratory Standards Institute (CLSI) recommends that clinical laboratories test all isolates of S. pneumoniae from CSF for resistance to penicillin, cefotaxime or ceftriaxone, meropenem, and vancomycin.[85] For organisms from other sources, laboratories should consider testing for resistance to erythromycin, penicillin, trimethoprim-sulfamethoxazole, clindamycin, cefepime, cefotaxime or ceftriaxone, a fluoroquinolone, meropenem, tetracycline, and vancomycin. Pneumococci resistant to vancomycin have never been described. Linezolid-resistance is extremely rare and has been associated with mutations within the rplD-encoded ribosomal protein L4.[86] For vancomycin, a strain is considered non-susceptible if it has a minimum inhibitory concentration of >1 μg/ml or greater or zone diameter less than <17 mm. For linezolid, nonsusceptible strains are those with a minimum inhibitory concentration (MIC) of >2 μg/ml or zone diameter <21 mm. Strains found to be nonsusceptible to vancomycin or linezolid should be submitted to a reference laboratory for confirmatory testing, and if resistant, reported to the state health department. Because pneumococci are fastidious organisms, some susceptibility testing methods used for other organisms are not appropriate for pneumococci; see the CLSI document for testing recommendations.[85] The CDC’s Antibiotic Resistance Laboratory Network (ARLN) is available to assist state public health laboratories with susceptibility testing of IPD isolates. States can request testing for select IPD isolates. Contact ARLN@cdc.gov for information on this program. Additionally, isolates with unusual resistance features can be sent to the CDC Streptococcus Laboratory for phenotypic verification and genomic analysis employing their specialized bioinformatics pipeline for detecting resistance determinants.[87] Contact pneumococcus@cdc.gov for additional information. State health department laboratories that obtain penicillin binding protein gene sequencing information may consider using the CDC Streptococcus Laboratory database for deducing β-lactam antibiotic MICs.[88]
Serotyping
Current pneumococcal vaccines are based upon capsular polysaccharides. There are currently >91 known capsular serotypes. Since only subsets of capsular serotypes are included in pneumococcal vaccines, serotyping allows the measurement of vaccine efficacy and can provide data for development of expanded-serotype vaccines.[89] CDC and its partners perform active, population-based surveillance for invasive pneumococcal serotypes in specific areas that represent about 30 million people in the United States. Serotyping is currently performed in only a limited number of state public health laboratories, academic centers, or at CDC. State public health laboratories may consider adopting a PCR-based technique for determining capsular serotypes.[90-93] The CDC Streptococcus Laboratory provides numerous protocols and references for state health departments and clinical labs to identify pneumococcal serotypes using PCR. If states are unable to perform PCR serotyping, the CDC’s ARLN and VPD programs can provide serotyping assistance for select IPD isolates or specimens. Contact ARLN@cdc.gov for serotyping assistance. The CDC Streptococcus Laboratory will conduct serotyping of pneumococcal isolates from blood, CSF, or other sterile sites in outbreak settings, and when appropriate will perform whole genome sequence analysis to determine strain features and relatedness between isolates from disease clusters. Contact pneumococcus@cdc.gov for outbreak assistance. Serotypes are deduced through the same bioinformatics pipeline used to determine MLST types and resistance features.[87.]
K. Poliomyelitis (see Chapter 12)
Virus detection
The likelihood of poliovirus isolation is highest from stool specimens, intermediate from pharyngeal swabs, and very low from blood or spinal fluid. Poliovirus is present in the stool in the highest concentration and for the longest time of any specimen, and therefore remains the most critical specimen for diagnosis. Enterovirus molecular typing, which includes PCR plus sequencing of the VP1 genome region, is extremely sensitive for the detection of poliovirus. Detection of virus from CSF is diagnostic but is rarely accomplished. To increase the probability of poliovirus isolation, at least two stool specimens and two throat swabs should be obtained 24 hours apart from patients with suspected poliomyelitis as early in the course of the disease as possible (i.e., immediately after poliomyelitis is considered as a possible differential diagnosis), but ideally within the first 14 days after onset of paralytic disease. Specimens should be collected and sent to the CDC polio laboratory for confirmation and to differentiate between possible wild-type or vaccine-derived strains[94].
Detection of wild poliovirus constitutes a public health emergency, and appropriate control efforts must be immediately initiated (in consultation among healthcare providers, the state and local health departments, and CDC). Wild poliovirus type 2 was declared eradicated in 2015, followed by wild poliovirus type 3 in 2019; wild and VDPV type 2 and 3 infectious and potentially infectious materials should be handled only in a proper containment facility.
L. Rotavirus (see Chapter 13)
Laboratory testing is necessary to confirm group A rotavirus infection and to ensure reliable surveillance and clinical therapy. Because rotavirus is shed in such high concentrations in stool, fecal specimens are preferred for diagnosis of rotavirus. Methods available to diagnose rotavirus infection include detection of viral antigens (EIA, immunochromatography, electron microscopy, and immunostaining) and molecular detection by RT-PCR/qRT-PCR and genomic sequencing.[95-97]
Detection of viral antigens
The most widely available method of antigen detection in stool is EIA, which detects an antigen common to all group A rotaviruses.[95] Several inexpensive commercial EIA kits are available and provide rapid and highly sensitive results (90%-100%). Because EIA is rapid, inexpensive, and highly sensitive, it is the most appropriate method for clinical diagnosis and surveillance.
Another less frequently used method more appropriate for a research setting is visualization of viral particles by electron microscopy.
Molecular detection
Molecular methods can be used to detect rotavirus infection in clinical specimens and to characterize the virus. Molecular methods for detection of viral RNA include RT-PCR, qRT-PCR, and genomic sequencing.[96,97]
- Multiplexed, semi-nested RT-PCR genotyping and genomic sequencing are widely used to identify the most common and several uncommon rotavirus G and P genotypes.[96,97]
- Genomic sequencing has been used extensively to identify uncommon strains and genetic variants that cannot be identified by RT-PCR genotyping and to confirm the results of genotyping methods.[96,97]
Virus isolation
Rotavirus can be isolated directly from fecal specimens by inoculation of cell cultures in the presence of trypsin-containing growth medium, but this procedure is labor intensive and more appropriate for research laboratories.
M. Rubella (see Chapter 14)
Clinical diagnosis of rubella is unreliable, therefore, cases must be laboratory confirmed. Virus detection and serologic testing can be used to confirm acute or recent rubella infection. Serologic tests can also be used to screen for rubella immunity.
Virus detection (real-time RT-PCR, RT-PCR)
Rubella virus can be detected from nasal, throat, urine, blood, and CSF specimens from persons with rubella (see Appendix 15 [2 pages]. The best results come from throat swabs. CSF specimens should be reserved for persons with suspected rubella encephalitis. Efforts should be made to obtain clinical specimens for virus detection from all case-patients at the time of the initial investigation. Virus may be detected from 1 week before to 2 weeks after rash onset. However, maximum viral shedding occurs up to day 4 after rash onset.
Real-time RT-PCR and RT-PCR can be used to detect rubella virus and has been extensively evaluated for its usefulness in detecting rubella virus in clinical specimens.[84,85] Clinical specimens obtained for virus detection and sent to CDC are routinely screened by these techniques.
Molecular typing is recommended because it provides important epidemiologic information to track the epidemiology of rubella in the United States now that rubella virus no longer continuously circulates in this country. By comparing virus sequences obtained from new case-patients with other virus sequences, the origin of particular virus types in this country can be tracked.[100] Furthermore, this information may help in documenting the maintenance of the elimination of endemic transmission. In addition, genotyping methods are available to distinguish wild-type rubella virus from vaccine virus.
Serologic testing
The serologic tests available for laboratory confirmation of rubella infections and immunity vary among laboratories. The state health department can provide guidance on available laboratory services and preferred tests.
EIAs are the most commonly used and widely available diagnostic test for rubella IgG and IgM antibodies and are sensitive and relatively easy to perform. EIA is the preferred testing method for IgM, using the capture technique, although indirect assays are also acceptable.
Latex agglutination tests appear to be sensitive and specific for screening when performed by experienced laboratory personnel. Other tests in limited use to detect rubella-specific IgM include HI and IFA.
Detection of IgM antibody
Rubella-specific IgM can usually be detected 4–30 days after onset of illness, and often for longer. Sera should be collected as early as possible after onset of illness. However, IgM antibodies may not be detectable before day 5 after rash onset. In case of a rubella IgM-negative result in specimens taken before day 5, serologic testing should be repeated on a specimen collected after day 5.
Because rubella incidence is low, a high proportion of IgM-positive tests will likely be false positive. False-positive serum rubella IgM tests may occur due to the presence of rheumatoid factors (indicating rheumatologic disease) or cross-reacting IgM, or infection with other viruses.[101,102] Avidity testing (see below) and detection of wild-type rubella virus can be used to resolve uncertainties in the serologic evaluation of suspected cases.
Particular care should be taken when rubella IgM is detected in a pregnant woman with no history of illness or contact with a rubella-like illness. Although this is not recommended, many pregnant women with no known exposure to rubella are tested for rubella IgM as part of their prenatal care. If rubella test results are IgM-positive for persons who have no or low risk of exposure to rubella, additional laboratory evaluation should be conducted. Laboratory evaluation is similar to that described in the IgM-positive section of Figure 2.
Detection of IgG antibody (significant rise or avidity) for diagnostic testing
To detect a significant rise in rubella-specific IgG concentration, the first serum specimen should be obtained as soon as possible after onset of illness and the second serum sample should be collected about 7–21 days after the first specimen. In most rubella cases, rubella IgG is detectable by 8 days after rash onset.[103] Tests for IgG antibody should be conducted on both acute-and convalescent-phase specimens at the same time with the same test.
Assays for IgG avidity are useful to distinguish the difference between recent and past rubella infections. Low avidity is associated with recent primary rubella infection, whereas high avidity is associated with past infection or reinfection. Avidity tests are not routine tests and should be performed in reference laboratories. A number of avidity assays have been described.[104,105]
Detection of IgG antibody to screen for rubella immunity
A single serologic IgG test may be used to determine the rubella immune status of persons whose history of rubella disease or vaccination is unknown. The presence of serum IgG rubella-specific antibodies indicates immunity to rubella.
N. Congenital Rubella Syndrome (see Chapter 15)
Diagnostic tests used to confirm CRS include serologic assays and detection of the virus.
Virus detection (real-time RT PCR and RT-PCR)
Rubella virus can be detected from nasal, throat, urine, and blood specimens from infants with CRS. Efforts should be made to obtain clinical specimens for virus isolation from infants at the time of the initial investigation (see Appendix 15 [2 pages]). However, because infants with CRS may shed virus from the throat and urine for a prolonged period (a year or longer), specimens obtained later may also yield rubella virus. As with rubella infection, molecular typing is recommended because it provides important epidemiologic information to track the epidemiology of rubella in the United States now that rubella virus no longer continuously circulates in this country. By comparing virus sequences from new case-patients with virus sequences from other cases, the origin of particular virus types in this country can be tracked.[106] Furthermore, this information may help in documenting the maintenance of the elimination of endemic rubella virus transmission. Specimens for molecular typing should be obtained from patients with CRS as soon as possible after diagnosis. Appropriate specimens include throat swabs, urine, and cataracts from surgery. Specimens for virus detection and molecular typing should be sent to CDC as directed by the state health department.
Serologic testing
The serologic tests available for laboratory confirmation of CRS infections vary among laboratories. EIAs are the most commonly used and widely available diagnostic test for rubella IgG and IgM antibodies. EIAs are sensitive and relatively easy to perform. EIA is the preferred testing method for IgM, using the capture technique, although indirect assays are also acceptable. In infants with CRS, IgM antibody can be detected in the infant’s cord blood or serum and persists for about 6–12 months. Suspect CRS cases that are IgM negative at birth should have this result confirmed at one month of age.
O. Varicella (see Chapter 17)
Laboratory confirmation of varicella has become more important due to the common vaccine-modified presentation of the disease and diminishing experience of physicians with unmodified disease. In addition, PCR testing that discriminates vaccine from wildtype varicella zoster virus (VZV) is important for the confirmation of vaccine-associated disease. Since varicella is the most common disease confused with orthopox disease (e.g.,mpox, smallpox), rapid laboratory confirmation of varicella is required in cases of vesicular/pustular rash illness that fall into the category of “moderate risk” for orthopox disease, according to the CDC differential algorithm. Diagnostic tests used to confirm recent varicella infection include PCR, serology, DFA, and culture. Serologic tests, DFA, and culture have limited value in confirming varicella etiology but are still in use.
Rapid varicella zoster virus identification
Skin lesions are the preferred sample for laboratory confirmation of varicella. Peripheral blood samples (serum or plasma) are used to test for varicella immunity. Samples from skin lesions should be collected by unroofing a vesicle, preferably a fluid-filled vesicle, then rubbing the base of the skin lesion with a synthetic swab. Collecting skin lesion specimens from breakthrough cases can be challenging because the rash is often maculopapular with few or no vesicles. If only macules or papules are present, suitable samples can be obtained by scraping the lesion (e.g., with the edge of a glass microscope slide), swabbing the abraded lesion, and then collecting any material on the object that was used to scrape the lesion on the same swab. Scabs from skin lesions are also excellent samples for detection of VZV DNA. Other sources such as nasopharyngeal secretions, saliva, blood, or urine are not recommended as they are less sensitive than swabs and scabs from skin lesions and often lead to false negative results. Viral DNA may also be detected in CSF in cases of neurologic disease and vasculopathies associated with VZV infection, or in bronchial washings/bronchoalveolar lavage in VZV-associated pneumonia. A video demonstrating the techniques for collecting various specimens for varicella confirmation, including specimens from breakthrough cases, can be found on the CDC varicella web site. Additional information about collecting and submitting specimens for testing can also be found on the CDC varicella web site or by contacting rbeard@cdc.gov.
PCR is the gold standard for confirming VZV infection, being sensitive, specific, and widely available. Short turnaround times of several hours are possible. Other direct detection methods such as DFA are generally not recommended due to limitations in both sensitivity and specificity. Suitable samples for PCR testing include vesicular swabs, scabs from crusted lesions, and biopsy or autopsy samples from cases of suspected disseminated VZV disease. Viral DNA may also be detected in CSF in cases of neurologic disease and vasculopathies associated with VZV infection. If PCR results on CSF are negative, a VZV IgG intrathecal antibody assay may yield a positive result to implicate VZV as much as one month after disease onset. Viral DNA is less frequently detected if more than 10 days post-onset of symptoms.
Postvaccination situations for which specimens should be tested include: 1) rash with more than 50 lesions occurring 7 or more days after vaccination, 2) suspected secondary transmission of the vaccine virus, 3) herpes zoster in a vaccinated person, or 4) any serious adverse event. Methods are available in specialized laboratories to identify VZV strains and to distinguish wild-type VZV from the vaccine (Oka/Merck) strain. Such testing is used in situations when it is important to distinguish wild-type from vaccine-type virus, e.g., in suspected vaccine adverse events. The National VZV Laboratory at CDC and the American Public Health Laboratory Association Vaccine Preventable Diseases Reference Centers (VPD-RCs [2 pages]) have the capacity to distinguish wild-type VZV from the vaccine strains using real-time PCR. VPD-RCs are located in the state laboratories of Wisconsin, California, New York, and Minnesota, and each VPD-RC receives specimens from a designated group of states.
Virus culture
The diagnosis of VZV infection may be confirmed by culture (isolation) of VZV but is generally not recommended because of the length of time for results and the insensitivity of the method compared to PCR. Although newer, rapid culture techniques can provide results within 2–3 days, they are still substantially less sensitive than PCR and not offered by most labs.
Serologic testing
Serologic tests are available for IgG (acute and convalescent) and IgM antibodies to VZV for confirmation of disease. However, paired IgG acute- and convalescent-phase antibody tests are not practical for immediate clinical management, and in vaccinated persons, a 4-fold rise may not occur. Testing using commercial kits for IgM antibody is not recommended since available methods lack sensitivity and specificity; false-positive IgM results are especially common in the presence of high IgG levels. Although not widely available, VZV IgG avidity can distinguish between primary VZV infection and past infection.
References
A. Diphtheria
- World Health Organization. (2021). WHO laboratory manual for the diagnosis of diphtheria and other related infections. World Health Organization. https://apps.who.int/iris/handle/10665/352275. License: CC BY-NC-SA 3.0 IGO
- Efstratiou A, Engler KH, Mazurova IK et al. Current approaches to the laboratory diagnosis of diphtheria. J Infect Dis 2000;181(Suppl 1):S138-45. doi: 10.1086/315552
- Engler KH, Glushkevich T, Mazurova IK et al. A modified Elek test for the detection of corynebacteria in the diagnostic laboratory. J Clin Microbiol 1997;35:495-98. doi: 10.1128/jcm.35.2.495-498.1997
- Williams MM, Waller JL, Aneke JS, et al. Detection and characterization of diphtheria toxin gene-bearing Corynebacterium species through a new real-time PCR assay. J Clin Microbiol 2020;58: doi: 10.1128/JCM.00639-20.Print 2020 Sep 22
- Dingle TC, Butler-Wu SM. Maldi-tof mass spectrometry for microorganism identification. Clin Lab Med 2013;33(3):589-609. doi: 10.1016/j.cll.2013.03.001
B. Hib
- Wang X, Mair R, Hatcher C, et al. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. IJMM 2010;301(4): 303–9. doi:10.1016/j.ijmm.2010.11.004
- Vuong J, Collard JM, Whaley MJ, et al. Development of real-time PCR methods for the detection of bacterial meningitis pathogens without DNA extraction. PLoS One 2016;11(2):e0147765. doi:10.1371/journal.pone.0147765
- Wang X, Theodore MJ, Mair R, et al. Clinical validation of multiplex real-time PCR assays for detection of bacterial meningitis pathogens. J Clin Microbiol 2012;50(3):702–8. doi:10.1128/ JCM.06087-11
- Leber AL, Everhart K, Balada-Llasat J-M, et al. Multicenter evaluation of BioFire FilmArray Meningitis/Encephalitis Panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol 2016;54(9):2251–61. doi:10.1128/JCM.00730-16
- Altun O, Almuhayawi M, Ullberg M, Özenci V. Rapid identification of microorganisms from sterile body fluids by use of FilmArray. J Clin Microbiol 2015;53(2):710–2. doi:10.1128/ JCM.03434-14
- CSTE. Vaccine preventable disease surveillance and reporting. CSTE position statement 09-ID-99. Atlanta, GA: CSTE; 2000.
- CDC. Serotyping discrepancies in Haemophilus influenzae type b disease—United States, 1998–1999. MMWR 2002;51(32):706–7.
- CDC. Best practices for use of PCR for diagnosing Haemophilus influenzae and Neisseria meningitidis and public health importance of identifying serotype/serogroup.
- Fry AM, Lurie P, Gidley M, et al. Haemophilus influenzae type b disease among Amish children in Pennsylvania: reasons for persistent disease. Pediatrics 2001;108(4):E60. doi:10.1542/peds.108.4.e60
- Lucher LA, Reeves M, Hennessy T, et al. Reemergence, in southwestern Alaska, of invasive Haemophilus influenzae type b disease due to strains indistinguishable from those isolated from vaccinated children. J Infect Dis 2002;186:958–65. doi:10.1086/342595
C. HAV
- HAV. Vaughan G, Goncalves Rossi LM, Forbi JC, et al. Hepatitis A virus: host interactions, molecular epidemiology and evolution. Infect Genet Evol 2014;21:227–43.
D. HBV
- CDC. Prevention of hepatitis B virus infection in the United States: recommendations of the Advisory Committee on Immunization Practices. MMWR Recomm and Rep 2018;67(RR-1):1–31.
- Mast EE, Margolis HS, Fiore AE, et al. Advisory Committee on Immunization Practices (ACIP). A comprehensive immunization strategy to eliminate transmission of hepatitis B virus infection in the United States: recommendations of the Advisory Committee on Immunization Practices (ACIP). Part 1: immunization of infants, children, and adolescents. MMWR Recomm Rep 2005;54(RR-16):1–31.
- Yang HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med 2002;347:168–74.
- Ott JJ, Stevens GA, Wiersma ST. The risk of perinatal hepatitis B virus transmission: hepatitis B e antigen (HBeAg) prevalence estimates for all world regions. BMC Infect Dis 2012;12:131. doi: 10.1186/1471-2334-12-131
- Forbi J, Vaughan G, Purdy M, et al. Epidemic history and evolutionary dynamics of hepatitis B virus infection in two remote communities in rural Nigeria. PLoS One 2010;5: e1615. doi: 10.1371/journal.pone.0011615
- Locarnini SA, Yuen L. Molecular genesis of drug-resistant and vaccine escape HBV mutants. Antivir Ther 2010;14(3 Pt b):451–61.
- Zhang AY, Lai CL, Huang FY, et al. Deep sequencing analysis of quasispecies in the HBV pre-S region and its association with hepatocellular carcinoma. J Gastroenterology 2017;52:1064–74.
- Longmire AG, Sims S, Rytsareva I, et al. GHOST: global hepatitis outbreak and surveillance technology. BMC Genomics 2017;18(Suppl 10):916. doi: 10.1186/s12864-017-4268-3
- Di Lello FA, Ridruejo E, Martinez AP, Perez PS, Campos RH, Flichman DM. Molecular epidemiology of hepatitis B virus mutants associated with vaccine escape, drug resistance and diagnosis failure. J Vir Hepatitis 2019;26:552–60.
- Thomas HC, Carman WF. Envelope and precore/core variants of hepatitis B virus. Gastroenterol Clin North Am 1994;23:499–514. https://pubmed.ncbi.nlm.nih.gov/7989091/
E. Influenza
- Bellini WJ, Cox NJ. Laboratory diagnosis of measles, influenza and other respiratory virus infections. Course manual. Washington, DC: HHS, Public Health Service; 1995.
- Ziegler T, Cox NJ. Influenza viruses. In: Murray PR, Barron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. Washington, DC: ASM Press; 1999.
- CDC. Rapid influenza diagnostic tests. Atlanta, GA [reviewed 2016 October 25; updated 2017 January 18; cited 2017 September 7].
- CDC. Evaluation of rapid influenza diagnostic tests for detection of novel influenza A(H1N1)— United States, 2009. MMWR Morb Mortal Wkly Rep 2009;58(30):826–9.
- CDC. Evaluation of 11 commercially available rapid influenza diagnostic tests—United States, 2011–2012. MMWR Morb Mortal Wkly Rep 2012;61(43):873–6.
- Moesker FM, van Kampen JJA, Aron G, et al. Diagnostic performance of influenza viruses and RSV rapid antigen detection tests in children in tertiary care. J Clin Vir 2016;79:12–7. doi: 10.1016/j. jcv.2016.03.022
F. Measles
- Jenkerson SA, Beller M, Middaugh JP, Erdman DD. False-positive rubeola IgM tests. N Engl J Med 1995;332:1103–4. doi:10.1056/NEJM199504203321616
- Helfand RF, Heath JL, Anderson LJ, Maes EF, Guris D, Bellini WJ. Diagnosis of measles with an IgM capture EIA: the optimal timing of specimen collection after rash onset. J Infect Dis 1997;175:195–9. doi:10.1093/infdis/175.1.195
G. Meningococcal Disease
- Rosenstein NE, Perkins BA, Stephens DS, et al. Meningococcal disease. N Engl J Med 2001;344:1378–88. doi: 10.1056/NEJM200105033441807
- Rouphael NG, Stephens DS. Neisseria meningitidis: Biology, microbiology, and epidemiology. Methods Mol Biol 2012;799:1–20. doi:10.1007/978-1-61779-346-2_1
- Mothershed EA, Sacchi CT, Whitney AM, et al. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol 2004;42:320–8. doi:10.1128/jcm.42.1.320-328.2004
- Biofire FilmArray® Panels.
- ARUP Meningitis/Encephalitis Panel by PCR.
- American Academy of Pediatrics. Meningococcal infections. In: Pickering LK, Baker CJ, Long SS, McMillan JA, editors. Red book: 2006 report of the committee on infectious diseases. 27th ed. Elk Grove Village, IL: American Academy of Pediatrics; 2006. 452–60.
- Raghunathan PL, Bernhardt SA, Rosenstein NE. Opportunities for control of meningococcal disease in the United States. Annu Rev Med 2004;55:333–53.
doi: 10.1146/annurev.med.55.091902.103612 - CDC. Prevention and control of meningococcal disease. MMWR Recomm Rep 2005;54(No. RR-7):1–17.
- Sacchi, CT, Whitney AM, Reeves MW, Mayer LW, Popovic T. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J Clin Microbiol 2002;40(12):4520–7. doi:10.1128/jcm.40.12.4520-4527.2002
- Maiden, MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA 1998;95:3140–5.
H. Mumps
- Bitsko RH, Cortese MM, Dayan GH, et al. Detection of RNA of mumps virus during an outbreak in a population with a high level of measles, mumps, and rubella vaccine coverage. J Clin Microbiol2008;46(3):1101–3. doi:1128/JCM.01803-07
- Rota JS, Rosen JB, Doll MK, et al. Comparison of the sensitivity of laboratory diagnostic methods from a well-characterized outbreak of mumps in New York City, 2009. Clin Vaccine Immunol 2013; 20(3):391–6. doi:10.1128/CVI.00660-12
- Rota JS, Turner JC, Yost-Daljev MK, et al. Investigation of a mumps outbreak among university students with two measles-mumps-rubella (MMR) vaccinations, Virginia, September-December 2006. J Med Virol 2009;81(10):1819–25. doi:10.1002/jmv.21557
- Nunn A, Masud S, Krajden M, et al. Diagnostic yield of laboratory methods and value of viral genotyping during an outbreak of mumps in a partially vaccinated population in British Columbia, Canada. J Clin Microbiol 2018;56(5):e01954-17. doi:10.1128/JCM.01954-17
- Fanoya EB, Cremer J, Ferreira JA, et al. Transmission of mumps virus from mumps-vaccinated individuals to close contacts. Vaccine 2011;29 (51):9551–6. doi:10.1016/j.vaccine.2011.09.100
- Cunningham C, Faherty C, Cormican M, Murphy AW. Importance of clinical features in diagnosis of mumps during a community outbreak. Ir Med J 2006;99(6):171–3.
- Krause CH, Molyneaux PJ, Ho-Yen DO, et al. Comparison of mumps-IgM ELISAs in acute infection. J Clin Virol 2007;38(2):153–6. doi:10.1016/j.jcv.2006.10.010
- Jin L, Feng Y, Parry R, et al. Real-time PCR and its application to mumps rapid diagnosis. J Med Virol 2007;79(11):1761–7. doi:10.1002/jmv.20880
- Tan KE, Anderson M, Krajden M, et al. Mumps virus detection during an outbreak in a highly unvaccinated population in British Columbia. Can J Public Health 2011;102(1):47–50. doi:10.1007/BF03404876
- Maillet M, Bouvat E, Robert N, et al. Mumps outbreak and laboratory diagnosis. J Clin Virol 2015;62:14–9. doi:10.1016/j.jcv.2014.11.004
- D’Huillier AG, Eshaghi A, Racey CS, et al. Laboratory testing and phylogenetic analysis during a mumps outbreak in Ontario, Canada. Virol J 2018;15(1):98. doi:10.1186/s12985-018-0996-5
- Krause CH, Eastick K, Ogilvie MM. Real-time PCR for mumps diagnosis on clinical specimens—comparison with results of conventional methods of virus detection and nested PCR. J Clin Virol 2006;37(3):184–9. doi: 10.1016/j.jcv.2006.07.009
- Hatchette T, Davidson R, Clay S, et al. Laboratory diagnosis of mumps in a partially immunized population: The Nova Scotia experience. Can J Infect Dis Med Microbiol 2009;20(4):e157–62. doi:10.1155/2009/493275
- Jin L, Orvell C, Myers R, et al. Genomic diversity of mumps virus and global distribution of the 12 genotypes. Rev Med Virol 2015;25(2):85–101. doi:10.1002/rmv.1819
- Wohl S, Metsky HC, Schaffner SF, et al. Combining genomics and epidemiology to track mumps virus transmission in the United States. PLoS Biol 2020;18(2):e3000611. doi:10.1371/journal.pbio.3000611
- McNall R, Wharton A, Anderson R, et al. Genetic Characterization of mumps viruses associated with the resurgence of mumps in the United States: 2015—2017. Virus Res 2020;16:197935. doi:10.1016/j.virusres.2020.197935
- Ukkonen P, Granström ML, Penttinen K. Mumps-specific immunoglobulin M and G antibodies in natural mumps infection as measured by enzyme-linked immunosorbent assay. J Med Virol 1981;8:131–42. doi: 10.1002/jmv.1890080207
- Benito RJ, Larrad L, Lasierra MP, et al. Persistence of specific IgM antibodies after natural mumps infection. J Infect Dis 1987;155:156–7. doi:10.1093/infdis/155.1.156
I. Pertussis
- Bejuk D, Begovac J, Bace A, et al. Culture of Bordetella pertussis from three upper respiratory tract specimens. Pediatr Infect Dis J 1995;14:64–5.
- Halperin SA, Bortolussi R, Wort AJ. Evaluation of culture, immunofluorescence, and serology for the diagnosis of pertussis. J Clin Microbiol 1989;27:752–7.
- Hallander HO, Reizenstein E, Renemar B, et al. Comparison of nasopharyngeal aspirates with swabs for culture of Bordetella pertussis. J Clin Microbiol 1993;31:50–2.
- Hoppe JE, Weiss A. Recovery of Bordetella pertussis from four kinds of swabs. Eur J Clin Microbiol 1987;6:203–5.
- Koidl C, Bozic M, Burmeister A, et al. Detection and differentiation of Bordetella spp. by real-time PCR. J Clin Microbiol 2007;45(2):347–50. doi:10.1128/JCM.01303-06
- Qin X, Galanakis E, Martin ET, Englund JA. Multi-target polymerase chain reaction for diagnosis of pertussis and its clinical implications. J Clin Microbiol 2007;45(2):506–11. doi:10.1128/JCM.02042-06
- Meade BD, Bollen A. Recommendations for use of the polymerase chain reaction in the diagnosis of Bordetella pertussis. J Med Microbiol 1994;41:51–5.
- Mandal S, Tatti KM, Woods-Stout D, et al. Pertussis pseudo-outbreak linked to specimens contaminated by Bordetella pertussis DNA from clinic surfaces. Pediatrics 2012;129:e424–30. doi:10.1542/peds.2011-1710
- Faulkner A, Skoff TH, Tondella L, et al. Trends in pertussis diagnostic testing in the United States, 1990—2012. Pediatr Infect Dis J 2016;35(1)39–44. doi:10.1097/INF.0000000000000921
- CDC. Best practices for health care professionals on the use of polymerase chain reaction (PCR) for diagnosing pertussis. 2011. Atlanta, GA: CDC, 2011.
- Tatti KM, Sparks KN, Boney KO, Tondella ML. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol 2011;49:4059–66. doi:10.1128/JCM.00601-11
- Marchant CD, Loughlin AM, Lett SM, et al. Pertussis in Massachusetts, 1981—1991: incidence, serologic diagnosis, and vaccine effectiveness. J Infect Dis 1994;169:1297–1305.
- Menzies SL, Kadwad V, Pawloski LC, et al. Development and analytical validation of an immunoassay for quantifying serum anti-pertussis toxin antibodies resulting from Bordetella pertussis. Clin Vaccine Immunol 2009;16(12):1781–8. doi:10.1128/CVI.00248-09
- Baughman AL, Bisgard KM, Edwards KM, et al. Establishment of diagnostic cutoff points for levels of serum antibodies to pertussis toxin, filamentous hemagglutinin, and fimbriae in adolescents and adults in the United States. Clin Diagn Lab Immunol 2004;11(6):1045–53. doi:10.1128/CDLI.11.6.1045-1053.2004
- Burgos-Rivera B, Lee AD, Bowden KE, et al. Evaluation of level of agreement in Bordetella species identification in three U.S. laboratories during a period of increased pertussis. J Clin Microbiol 2015;53:1842–4. doi:10.1128/JCM.03567-14
- Schmidtke AJ, Boney KO, Martin SW, et al. Population diversity among Bordetella pertussis isolates, United States, 1935—2009. Emerg Infect Dis 2012;18:1248–55. doi:10.3201/eid1808.120082
- de Moissac YR, Ronald SL, Peppler MS. Use of pulsed-field gel electrophoresis for epidemiological study of Bordetella pertussis in a whooping cough outbreak. J Clin Microbiol 1994;32:398–402. doi:10.1128/JCM.32.2.398-402.1994
- Bisgard K, Christie C, Reising S, et al. Molecular epidemiology of Bordetella pertussis by pulsed-field gel electrophoresis profile: Cincinnati, 1989—1996. J Infect Dis 2001;183:1360–7. doi:10.1086/319858
- Schouls LM, van der Heide HG, Vauterin L, et al. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J Bacteriol 2004;186:5496–505. doi:10.1128/JB.186.16.5496-5505.2004
- Bowden KE, Williams MM, Cassiday PK, et al. Molecular epidemiology of the pertussis epidemic in Washington State in 2012. J Clin Microbiol 2014;52:3549–57. doi:10.1128/JCM.01189-14
- Litt DJ, Jauneikaite E, Tchipeva D, et al. Direct molecular typing of Bordetella pertussis from clinical specimens submitted for diagnostic quantitative (real-time) PCR. J Med Microbiol 2012;61:1662–8. doi: 10.1099/jmm.0.049585-0
- Maiden MC. Multilocus sequence typing of bacteria. Annu Rev of Microbiol 2006;60:561–88. doi:10.1146/annurev.micro.59.030804.121325
J. Pneumococcal (IPD)
- Clinical and Laboratory Standards Institute. M100-S22 Performance standards for antimicrobial susceptibility testing; Twenty-second informational supplement. Approved Standard. Wayne, PA: Clinical and Laboratory Standards Institute, 2012.
- Dong W, Chochua S, McGee L, Jackson D, Klugman KP, Vidal JE. Mutations within the rplD gene of linezolid-nonsusceptible Streptococcus pneumoniae strains isolated in the United States. Antimicrob Agents Chemother 2014;58(4):2459–62. doi:10.1128/AAC.02630-13
- Metcalf BJ, Chochua S, Gertz RE, Jr, Li Z, Walker H, Tran T, et al. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect 2016;22(12):1002 e1– e8. doi:10.1016/j.cmi.2016.08.001
- Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Jr., Walker H, et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting beta-lactam resistance levels in Streptococcus pneumoniae. MBio 2016;7(3) e00756–16. doi:10.1128/mBio.00756-16
- Beall B, McEllistrem MC, Gertz RE Jr, et al. Pre- and post-vaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. Active Bacterial Core Surveillance Team. J Clin Microbiol 2006;44(3):999–1017. doi:10.1128/JCM.44.3.999-1017.2006
- Pai R, Gertz RE, Beall B, et al. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae. J Clin Microbiol 2006;44(1):124–31. doi:10.1128/JCM.44.1.124-131.2006
- Carvalho MG, Tondella ML, McCaustland K, et al. Evaluation and Improvement of real-time PCR detection assays to lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 2007:45(8);2460–66. doi:1128/JCM.02498-06
- Dias CA, Teixeira LM, Carvalho MG, Beall B. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J Med Microbiol 2007;56(9):1185–8. doi:1099/jmm.0.47347-0
- Morais M, Carvalho MG, Roca, A, et al. Sequential multiplex PCR for identifying pneumococcal capsular serotypes from south-Saharan African clinical isolates. J Med Microbiol 2007;56:1181–4. doi:1099/jmm.0.47346-0
K. Polio
- Strebel PM, Sutter RW, Cochi SL, et al. Epidemiology of poliomyelitis in the United States one decade after the last reported case of indigenous wild virus-associated disease. Clin Infect Dis 1992;14:568–79. doi:10.1093/clinids/14.2.568
L. Rotavirus
- Gray JD, Desselberger U, editors. 2000. Rotavirus, methods & protocols. (Methods in molecular medicine). New York: Humana Press, 2000.
- Fischer TK, Gentsch JR. Rotavirus typing methods and algorithms. Rev Med Virol 2004;14:71–82. doi:10.1002/rmv.411
- Esona MD, Gautam R. Rotavirus. Diagnostic Testing for Enteric Pathogens. (Clinics in Laboratory Medicine). Philadelphia: Elsevier, 2015.
M. Rubella
- Namuwulya P, Abernathy E, Bukenya H, et al. Phylogenetic analysis of rubella viruses identified in Uganda, 2003-2012. J Med Virol 2014; 86(12):2107–13. doi:10.1002/jmv.23935
- Abernathy E, Cabezas C, Sun H, et al. Confirmation of rubella within 4 days of rash onset: comparison of rubella virus RNA detection in oral fluid with immunoglobulin M detection in serum or oral fluid. J Clin Microbiol 2009;47(1):182–8. doi:10.1128/JCM.01231-08
- Lazar M, Abernathy E, Chen M-H, Icenogle J, et al. Epidemiological and molecular investigation of a rubella outbreak, Romania, 2011 to 2012. Euro Surveill 2016;21(38):303–45. doi:10.2807/1560-7917
- Kurtz JB, Anderson MJ. Cross-reactions in rubella and parvovirus specific IgM tests. Lancet 1985;2:1356.
- Morgan-Capner P. False positive tests for rubella-specific IgM. Pediatr Infect Dis J 1991;10:415–6.
- Bellini WJ, Icenogle JP. Measles and rubella viruses. In: Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, and Warnock DW, editors. Manual of clinical microbiology, 11th ed. Washington, DC: American Society for Microbiology; 2015:1519–35.
- Hamkar R, Jalilvand S, Mokhtari-Azad T, et al. Assessment of IgM enzyme immunoassay and IgG avidity assay for distinguishing between primary and secondary immune response to rubella vaccine. J Virol Methods 2005;130(1–2):59–65. doi:10.1016/j.jviromet.2005.06.003
- Hofmann J, Liebert UG. Significance of avidity and immunoblot analysis for rubella IgM-positive serum samples in pregnant women. J Virol Methods 2005;130(1–2):66–71. doi:016/j.jviromet.2005.06.004
N. CRS
- Bosma TJ, Corbett KM, Eckstein MB, et al. PCR for detection of rubella virus RNA in clinical samples. J Clin Microbiol 1995;33:1075–9. doi:10.1128/JCM.33.5.1075-1079.1995
O. Varicella