Outbreak of Salmonella Newport Infections with Decreased Susceptibility to Azithromycin Linked to Beef Obtained in the United States and Soft Cheese Obtained in Mexico — United States, 2018–2019
Weekly / August 23, 2019 / 68(33);713–717
Please note: An erratum has been published for this report. To view the erratum, please click here.
Ian D. Plumb, MBBS1; Colin A. Schwensohn, MPH1; Laura Gieraltowski, PhD1; Selam Tecle, MPH2; Zachary D. Schneider, MPH1,3; Jennifer Freiman, MPH4; Andrea Cote, DVM4; Douglas Noveroske, MPH4; Jonathan Kolsin, MPH5; Joshua Brandenburg1,3; Jessica C. Chen, PhD1; Kaitlin A. Tagg, PhD1,6; Porscha Bumpus White, MS1; Hazel J. Shah, MPH1; Louise K. Francois Watkins, MD1; Matthew E. Wise, PhD1; Cindy R. Friedman, MD1 (View author affiliations)
View suggested citationSummary
What is already known about this topic?
Decreased susceptibility to azithromycin is rare among Salmonella serotypes that cause human infections in the United States. If antibiotic treatment is indicated, azithromycin is recommended as an oral therapy.
What is added by this report?
During June 2018–March 2019, an outbreak caused by multidrug-resistant Salmonella Newport with decreased susceptibility to azithromycin led to 255 infections and 60 hospitalizations. Infections were linked to Mexican-style soft cheese obtained in Mexico and beef obtained in the United States.
What are the implications for public health practice?
Whole genome sequencing can be used in Salmonella outbreak investigations for rapid prediction of antimicrobial resistance and can link cases to each other and to possible sources of infection.
Altmetric:
In September 2018, CDC identified Salmonella enterica serotype Newport (Newport) infections that were multidrug resistant (MDR), with decreased susceptibility to azithromycin, a recommended oral treatment agent. Until 2017, decreased susceptibility to azithromycin had occurred in fewer than 0.5% of Salmonella isolates from U.S. residents. This report summarizes the investigation of a multistate MDR Salmonella outbreak conducted by CDC, state and local health departments, and the U.S. Department of Agriculture’s Food Safety and Inspection Service. During June 2018–March 2019, 255 cases of infection with the outbreak strain were identified in 32 states; 43% of patients (89 of 206 with information on travel) reported recent travel to Mexico. Infections were linked to consumption of soft cheese obtained in Mexico and beef obtained in the United States. Consumers should avoid eating soft cheese that could be made from unpasteurized milk, regardless of the source of the cheese. When preparing beef, a food thermometer should be used to ensure that appropriate cooking temperatures are reached. When antibiotic treatment is needed for a patient, clinicians should choose antibiotics based on susceptibility testing wherever possible.
Epidemiologic Investigation
In 2018, during an investigation of antibiotic-susceptible Newport infections that led to a U.S. ground beef recall (1), a genetically distinct group of MDR Newport isolates was identified. Isolates were classified as the outbreak strain if they fell within the MDR clade (0–11 alleles by core genome multilocus sequence typing [cgMLST]); isolates were identified using PulseNet, the national subtyping network for foodborne bacterial disease surveillance. A case was defined as isolation of the outbreak strain from a patient during June 2018–March 2019. After interviews conducted by state and local health departments, some patients were reinterviewed using a standardized hypothesis-generating questionnaire or supplementary questionnaires that included questions about travel and antibiotic treatment. Food exposures were reported for the 7 days before illness onset. Exposures among patients who did not travel internationally were compared with those expected among a nationally representative sample of healthy persons included in the U.S. Foodborne Diseases Active Surveillance Network population survey (2006–2007) (2), after stratification by sex and ethnicity.
During June 2018–March 2019, 255 cases were identified in 32 U.S. states (Figure). Overall, 29% (60/209) of patients for whom information was available were hospitalized, 6% (4/70) were admitted to an intensive care unit, 4% (10/255) had Salmonella bacteremia, and two died. The median patient age was 36 years (range = <1–90 years), 58% (145/250) were female, and 65% (143/221) were Hispanic. Overall, 43% (89/206) with information on travel reported visiting Mexico in the 7 days preceding illness onset. Travelers to Mexico mostly reported visiting friends or family (67%, 24/36) and collectively reported visiting 16 of the 32 states within Mexico. Patients who did not visit Mexico were residents of 26 U.S. states.
Among patients who traveled to Mexico with information on food consumption, 87% (41/47) reported eating beef, and 63% (29/46) reported eating soft cheese; among those, 79% (23/29) recalled obtaining the cheese in Mexico (Table 1). Of several types of artisanal cheese reported, the most frequently recalled cheese was queso fresco, a cheese that is typically made with raw, unpasteurized milk from cows or goats (3). Among patients who did not travel to Mexico, 29% (20/70) reported eating Mexican-style soft cheese, and 93% (68/73) reported eating beef (Table 1). The percentage who ate Mexican-style soft cheese was similar to the percentage in the nationally representative sample of healthy persons (p-value = 0.54), whereas the percentage who ate beef was higher than that among healthy persons (p<0.01).
Product and Animal Testing
In September 2018, the outbreak strain was detected in a cecal sample from a steer collected at a slaughter and processing plant in Texas as part of National Antimicrobial Resistance Monitoring System (NARMS) surveillance (Figure). In October 2018, the outbreak strain was detected in a mixture of queso fresco and Oaxaca soft cheese purchased in a market in Tijuana, Mexico. The cheese had been brought into the United States by a patient who became ill with a strain that was indistinguishable (0 allele difference) from the strain isolated from the cheese. The outbreak strain was detected in beef samples collected in November 2018 and March 2019 at two Texas slaughter and processing facilities. Isolates from the Mexican cheese, the steer cecum, and beef differed by 0–2 alleles from one another and by a minimum of 0–1 alleles from patient isolates (Table 2). Review of patient information did not identify any common suppliers of contaminated beef or cheese.
Antibiotic Resistance
Antibiotic resistance was predicted using whole genome sequencing and confirmed in a subset of isolates by antimicrobial susceptibility testing using broth microdilution; decreased susceptibility to azithromycin was defined as minimum inhibitory concentration ≥32 μg/mL (4). Of 252 isolates with resistance information, 226 (90%) had predicted resistance to trimethoprim-sulfamethoxazole, tetracycline, and chloramphenicol, and decreased susceptibility to azithromycin. In 143 (57%) isolates, there was additional predicted resistance to ampicillin and streptomycin, and nonsusceptibility to ciprofloxacin (defined as minimum inhibitory concentration ≥0.12 μg/mL) (4). All resistance genes were located on an IncR plasmid. Among patients with treatment information, 65/87 (75%) received antibiotic therapy, and 28/86 (33%) received an antibiotic to which the outbreak strain was resistant or showed decreased susceptibility.
Discussion
This investigation identified an MDR strain of Salmonella Newport with decreased susceptibility to azithromycin and nonsusceptibility to ciprofloxacin, two oral agents recommended for treatment of Salmonella infections. The presence of resistance genes on a plasmid is concerning because of the potential for spread to other bacteria (5). The outbreak strain appears to have emerged recently because Newport with decreased susceptibility to azithromycin was not detected in animal, retail meat, or human isolates in NARMS surveillance before 2016 (4). During 2016–2017, two smaller multistate clusters of MDR Newport infections with decreased susceptibility to azithromycin were investigated among U.S. residents; isolates were within 11 alleles of the current outbreak isolates. No source of the infections was identified, but a high percentage of patients reported recent travel to Mexico (Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, CDC, unpublished data, 2019). Routine monitoring in 2016 detected an isolate from a sample of beef imported from Mexico that was indistinguishable (0 allele difference) from the outbreak strain isolated from cheese in 2018.
In this MDR outbreak, consumption of cheese and consumption of beef were both associated with illness, indicating that dairy cattle were a likely source of these infections. The detection of the outbreak strain in cheese purchased in Mexico and the high percentage of travelers to Mexico who reported eating Mexican-style soft cheese suggest that soft cheese from Mexico was a source of infection. Mexican-style soft cheese has been previously identified as a source of other Salmonella outbreaks (6). The reported consumption of queso fresco, travel to various regions in Mexico, and detection of indistinguishable Newport strains in beef and cheese suggest that contamination of soft cheese resulted from carriage by cattle rather than poor hygiene during cheese production. Dairy cattle often are used as a source of ground beef and have been implicated in previous MDR Newport outbreaks (5).
Among patients who did not travel to Mexico, beef was identified as a source of infection by the close genetic relatedness between isolates from patients and beef samples, and from the higher percentage of patients who ate beef compared with the percentage of healthy persons who ate beef. It is also possible that beef was a source of infection among some travelers to Mexico; nearly 90% of them also reported eating beef, and in 2016 the outbreak strain was detected in beef imported from Mexico.
The genetic similarity between isolates from beef in Mexico, beef in the United States, and a steer in the United States strongly suggests that the outbreak strain is present in cattle in both countries. Because use of antibiotics in livestock can cause selection of resistant strains (7), the reported 41% rise in macrolide use in U.S. cattle from 2016 to 2017 (8) might have accelerated carriage of the outbreak strain among U.S. cattle. Avoiding the unnecessary use of antibiotics in cattle, especially those that are important for the treatment of human infections, could help prevent the spread of MDR Newport with decreased susceptibility to azithromycin. Further investigation is warranted to determine the prevalence of Newport with decreased susceptibility to azithromycin in U.S. and Mexican cattle, and to identify measures to prevent transmission among cattle.
Whole genome sequencing was valuable in linking human infections to food sources, distinguishing the MDR outbreak strain from an antibiotic-susceptible strain causing a simultaneous outbreak, and predicting antibiotic resistance. In this outbreak, one in three patients received an antibiotic that was likely to have been ineffective. Clinicians should limit use of antibiotics for patients with an acute diarrheal illness to those with clinical indications (9), and antibiotic selection should be based on susceptibility results whenever possible. For empiric treatment of patients with suspected Newport with decreased susceptibility to azithromycin, ceftriaxone or alternative agents should be considered. To prevent infection, consumers should avoid eating soft cheese that could be made with unpasteurized milk, and when preparing beef they should use a thermometer to ensure appropriate cooking temperatures are reached: 145°F (62.8°C) for steaks and roasts followed by a 3-minute rest time, and 160°F (71.1°C) for ground beef or hamburgers (10).
Acknowledgments
Meseret Birhane, Katherine E. Heiman-Marshall, CDC; Sherry Shaw, Uday Dessai, Bonnie Kissler, Food Safety and Inspection Service, U.S. Department of Agriculture.
Corresponding author: Ian D. Plumb, iplumb@cdc.gov, 404-718-5543.
1Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, CDC; 2California Department of Public Health; 3Oak Ridge Institute for Science and Education, Tennessee; 4Food Safety and Inspection Service, U.S. Department of Agriculture, Washington, DC; 5Texas Department of State Health Services; 6Weems Design Studio Inc., Suwanee, Georgia.
All authors have completed and submitted the International Committee of Medical Editors form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
References
- CDC. Outbreak of Salmonella infections linked to ground beef. Food safety alert. Atlanta, GA: US Department of Health and Human Services, CDC; 2019. https://www.cdc.gov/salmonella/newport-10-18/index.html
- CDC. Foodborne Diseases Active Surveillance Network (FoodNet) population survey atlas of exposure, 2006–2007. Atlanta, GA: US Department of Health and Human Services, CDC; 2008. https://www.cdc.gov/foodnet/surveys/foodnetexposureatlas0607_508.pdf
- González-Córdova AF, Yescas C, Ortiz-Estrada ÁM, De la Rosa-Alcaraz MLÁ, Hernández-Mendoza A, Vallejo-Cordoba B. Invited review: artisanal Mexican cheeses. J Dairy Sci 2016;99:3250–62. CrossRef PubMed
- Food and Drug Administration, CDC, US Department of Agriculture. 2015 NARMS integrated report. Silver Spring, MD: US Department of Health and Human Services, Food and Drug Administration, CDC; US Department of Agriculture; 2015. https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2015-narms-integrated-report
- Varma JK, Marcus R, Stenzel SA, et al. Highly resistant Salmonella Newport-MDRAmpC transmitted through the domestic US food supply: a FoodNet case-control study of sporadic Salmonella Newport infections, 2002–2003. J Infect Dis 2006;194:222–30. CrossRef PubMed
- Gould LH, Mungai E, Behravesh CB. Outbreaks attributed to cheese: differences between outbreaks caused by unpasteurized and pasteurized dairy products, United States, 1998–2011. Foodborne Pathog Dis 2014;11:545–51. CrossRef PubMed
- Tang KL, Caffrey NP, Nóbrega DB, et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health 2017;1:e316–27. CrossRef PubMed
- Food and Drug Administration. 2017 summary report on antimicrobials sold or distributed for use in food-producing animals. Silver Spring, MD: US Department of Health and Human Services, Food and Drug Administration; 2018. https://www.fda.gov/media/119332/download
- Shane AL, Mody RK, Crump JA, et al. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 2017;65:e45–80. CrossRef PubMed
- US Department of Health and Human Services. FoodSafety.gov. Keep food safe: food safety by type of food. Washington DC: US Department of Health and Human Services; 2019. https://www.foodsafety.gov/keep/types/meat/index.html
 FIGURE. Identification of isolates of the outbreak strain of Salmonella enterica serotype Newport from infected patients (N = 255), by travel status,* and from nonhuman sources† (n = 4), by epidemiologic week and year — United States, June 2018–March 2019
FIGURE. Identification of isolates of the outbreak strain of Salmonella enterica serotype Newport from infected patients (N = 255), by travel status,* and from nonhuman sources† (n = 4), by epidemiologic week and year — United States, June 2018–March 2019
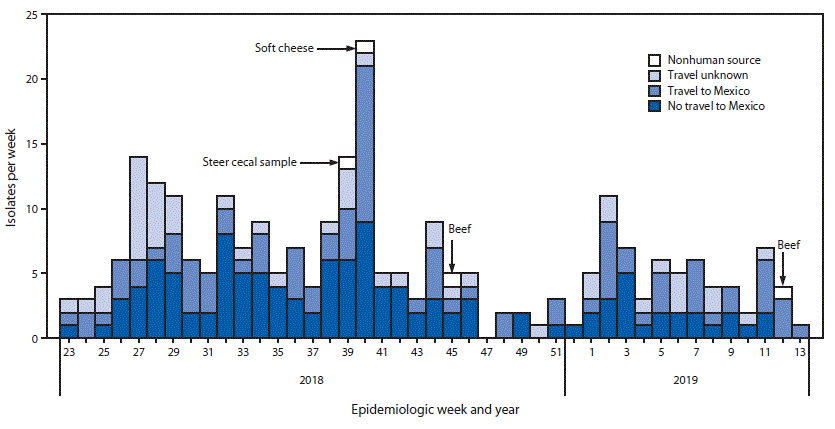
* Defined as reported travel within 7 days before illness onset.
† Cecal sample and beef samples obtained in the United States; sample of cheese obtained in Mexico by a patient infected with the outbreak strain who consumed this cheese.
* Of patients with known travel status, 89 had visited Mexico, and 117 had not visited Mexico.
† One patient who did not travel to Mexico reported eating beef obtained in Mexico in addition to beef obtained in the United States
§ One patient who traveled to Mexico reported eating Mexican-style soft cheese obtained in the United States and also Mexican-style soft cheese obtained in Mexico.
* Isolates were within 0–2 alleles of each other by core genome multilocus sequence typing.
† Obtained from the home of a patient who consumed some of it within 7 days before illness onset; the cheese was purchased from a market in Tijuana, Mexico.
Suggested citation for this article: Plumb ID, Schwensohn CA, Gieraltowski L, et al. Outbreak of Salmonella Newport Infections with Decreased Susceptibility to Azithromycin Linked to Beef Obtained in the United States and Soft Cheese Obtained in Mexico — United States, 2018–2019. MMWR Morb Mortal Wkly Rep 2019;68:713–717. DOI: http://dx.doi.org/10.15585/mmwr.mm6833a1.
MMWR and Morbidity and Mortality Weekly Report are service marks of the U.S. Department of Health and Human Services.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of
Health and Human Services.
References to non-CDC sites on the Internet are
provided as a service to MMWR readers and do not constitute or imply
endorsement of these organizations or their programs by CDC or the U.S.
Department of Health and Human Services. CDC is not responsible for the content
of pages found at these sites. URL addresses listed in MMWR were current as of
the date of publication.
All HTML versions of MMWR articles are generated from final proofs through an automated process. This conversion might result in character translation or format errors in the HTML version. Users are referred to the electronic PDF version (https://www.cdc.gov/mmwr) and/or the original MMWR paper copy for printable versions of official text, figures, and tables.
Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.