Multiple Locus Variable-number Tandem Repeat Analysis (MLVA)
What is MLVA (Multiple Locus Variable-Number Tandem Repeat Analysis)?
MLVA, an abbrevation for multiple locus variable number of tandem repeats analysis, is another technique used by microbiologists to generate a DNA fingerprint or a bacterial isolate. Microbiologists usually perform MLVA after they have performed Pulsed-Field Gel Electrophoresis (PFGE) so they can get more details about the type of bacteria that may be causing an outbreak.
MLVA Workflow: Larger View
Print version pdf icon[PDF – 1 page]
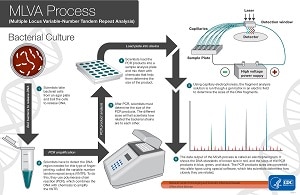
MLVA Process
- Scientists take bacterial cells from an agar plate and boil the cells to release DNA.
- Scientists have to detect the DNA region needed for this type of fingerprinting, called the variable-number tandem repeat arrays (VNTR). To do this, they use polymerase chain reaction (PCR), which combines the DNA with chemicals to amplify the VNTR.
- After PCR, scientists must determine the size of the PCR products. The different sizes will tell scientists how related the bacterial strains are to each other.
- Scientists load the PCR products into a sample analysis plate and mix them with chemicals that help them determine the size of the product.
- Using capillary electrophoresis, the fragment analysis solution is run though a gel matrix in an electric field to determine the sizes of the DNA fragments.
- The data output of the MLVA process is called an electropherogram. It shows the DNA standards of known size in red, and the sizes of the PCR products in blue, green, and black. The PCR products sizes are converted into allele types using special software, which lets scientists determine how closely they are related.
Advantages of MLVA
- MLVA may be able to differentiate suspected, fast-evolving bacterial strains from an outbreak even though those strains might look the same using other methods of DNA fingerprinting, such as PFGE..
- PulseNet uses MLVA as a complementary technique to PFGE, allowing microbiologists to see more detailed differences between bacteria that have similar PFGE patterns.
Limitations of MLVA
- Requires a trained and skilled technician
- Is not a practical routine subtyping method because a specific protocol must be used for each pathogen
- A few standardized protocols are available, but only the most common pathogens are subtyped by this method
MLVA Protocols
- Analysis: Shiga-Toxin Producing E. coli O157 Applied BioSystems Genetic Analyzer 3130/3500 pdf icon[PDF – 10 pages]
- Analysis: Shiga-Toxin Producing E. coli O157 Beckman Coulter CEQ 8000/8800/GeXP pdf icon[PDF – 10 pages]
- Analysis: Salmonella Enteritidis Beckman Coulter CEQ 8000/8800/GeXP pdf icon[PDF – 11 pages]
- Analysis: Salmonella Enteritidis Applied BioSystems Genetic Analyzer 3130/3500 pdf icon[PDF – 10 pages]
- Analysis: Salmonella Typhimurium Applied BioSystems Genetic Analyzer 3130/3500 pdf icon[PDF – 9 pages]
- Analysis: Salmonella Typhimurium Beckman Coulter CEQ 8000/8800/GeXP pdf icon[PDF – 10 pages]
- Laboratory Procedure: Shiga-Toxin Producing E. coli O157 Applied Biosystems Genetic Analyzer 3130XL pdf icon[PDF – 18 pages]
- Laboratory Procedure: Shiga-Toxin Producing E. coli O157 Applied Biosystems Genetic Analyzer 3500 Platform pdf icon[PDF – 20 pages]
- Laboratory Procedure: Salmonella Enteritidis Applied Biosystems Genetic Analyzer 3130XL Platform pdf icon[PDF – 18 pages]
- Laboratory Procedure: Salmonella Enteritidis Applied Biosystems Genetic Analyzer 3500 Platform pdf icon[PDF – 20 pages]
- Laboratory Procedure: Salmonella Enteritidis Beckman Coulter CEQ 8000/8800/GeXP Platform pdf icon[PDF – 17 pages]
- Laboratory Procedure: Salmonella Typhimurium Applied Biosystems Genetic Analyzer 3130XL Platform pdf icon[PDF – 18 pages]
- Laboratory Procedure: Salmonella Typhimurium Applied Biosystems Genetic Analyzer 3500 Platform pdf icon[PDF – 20 pages]
- Laboratory Procedure: Salmonella Typhimurium Beckman Coulter CEQ 8000/8800/GeXP Platform pdf icon[PDF – 17 pages]
Page last reviewed: February 16, 2016