Whole Genome Sequencing
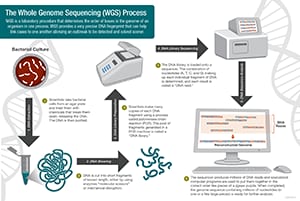
WGS Workflow: Larger View
Download [PDF – 1 page]
What is whole genome sequencing (WGS)?
All organisms (bacteria, vegetable, mammal) have a unique genetic code, or genome, that is composed of nucleotide bases (A, T, C, and G). If you know the sequence of the bases in an organism, you have identified its unique DNA fingerprint, or pattern. Determining the order of bases is called sequencing. Whole genome sequencing is a laboratory procedure that determines the order of bases in the genome of an organism in one process.
How does whole genome sequencing work?
Scientists conduct whole genome sequencing by following these four main steps:
- DNA shearing: Scientists begin by using molecular scissors to cut the DNA, which is composed of millions of bases (A’s, C’s, T’s and G’s), into pieces that are small enough for the sequencing machine to read.
- DNA bar coding: Scientists add small pieces of DNA tags, or bar codes, to identify which piece of sheared DNA belongs to which bacteria. This is similar to how a bar code identifies a product at a grocery store.
- DNA sequencing: The bar-coded DNA from multiple bacteria is combined and put in a DNA sequencer. The sequencer identifies the A’s, C’s, T’s, and G’s, or bases, that make up each bacterial sequence. The sequencer uses the bar code to keep track of which bases belong to which bacteria.
- Data analysis: Scientists use computer analysis tools to compare sequences from multiple bacteria and identify differences. The number of differences can tell the scientists how closely related the bacteria are, and how likely it is that they are part of the same outbreak.

How has whole genome sequencing improved disease detection?
Since 2019, whole genome sequencing has been the standard PulseNet method for detecting and investigating foodborne outbreaks associated with bacteria such as Campylobacter, Shiga toxin-producing E. coli (STEC), Salmonella, Vibrio, and Listeria. Since being launched, whole genome sequencing of pathogens in public health laboratories has improved surveillance for foodborne disease outbreaks and enhanced our ability to detect trends in foodborne infections and antimicrobial resistance. Whole genome sequencing provides detailed and precise data for identifying outbreaks sooner. Additionally, whole genome sequencing is used to characterize bacteria as well as track outbreaks; this greatly improves the efficiency of how PulseNet conducts surveillance.
PulseNet established the structure to support whole genome sequencing at state public health laboratories through:
- Training public health laboratory scientists to perform whole genome sequencing
- Purchasing equipment and supplies
- Updating data analysis systems and software
As the use of whole genome sequencing expands, CDC’s national surveillance systems and laboratory infrastructure must keep pace with the changing technology. With modernization, CDC and its public health partners can continue to successfully detect, respond to, and stop infectious diseases. Whole genome sequencing is a fast and affordable way to obtain detailed information about bacteria using just one test. Together, we can ensure rapid and less costly diagnoses for individuals and collect the evidence needed to quickly solve and prevent foodborne outbreaks.
The implementation of whole genome sequencing of pathogens for detecting and tracking foodborne outbreaks was made possible through collaborations with CDC’s Advanced Molecular Detection (AMD) Office, Food Safety Office, and Antimicrobial Resistance Solutions Initiative.
*For latest PulseNet laboratory protocols, please e-mail pulsenetngslab@cdc.gov