HCV Molecular Epidemiology
When someone is infected with the hepatitis C virus (HCV), the virus exists as a population of closely related, but not identical, variants. These variants are called quasispecies. Quasispecies continually evolve to form a unique “genetic fingerprint”. These genetic variations of HCV are useful for studying transmission links between patients, as one component of an epidemiologic investigation.1-4
Hepatitis C Virus (HCV) Phylogenetic Trees and GHOST reports
CDC’s Division of Viral Hepatitis’ molecular epidemiology laboratory uses a method called next generation sequencing to compare samples of hepatitis C virus (HCV) to see how closely the genetics of the virus match, and has developed and shared with specially-trained state public health laboratories a specific methodology called GHOST – Global Hepatitis Outbreak Surveillance Technology.2-4
This testing can help identify transmission linkages, however determining the source of infection or direction of transmission must be accomplished through other components of an epidemiologic investigation. For investigations involving persons who inject drugs, findings of no transmission linkage should be interpreted with caution because each time needles or other injection equipment are shared, new HCV strains can be introduced in the drug user which might obscure evidence of past linkages.
The test results, either in the form of a phylogenetic tree or GHOST diagram, help epidemiologists determine whether HCV infections share a common source of transmission. If samples have a very high degree of genetic relatedness (>96%) between their quasispecies of hepatitis C virus this means they are linked by transmission. 2-4
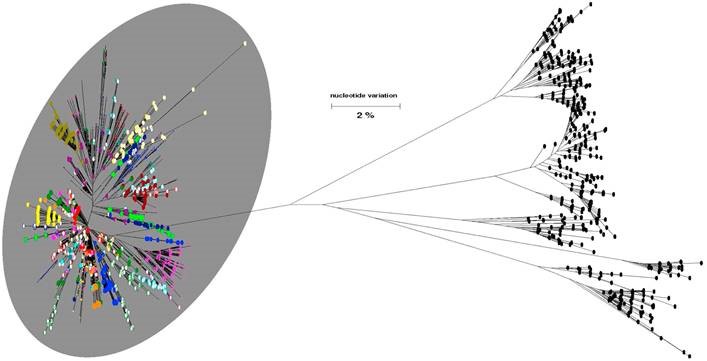 image iconexternal icon
image iconexternal icon
Sample phylogenetic tree – click on image for enlarged view
In phylogenetic tree reports each color (and randomly assigned case number if included) represents someone infected with hepatitis C virus. Multiple dots of the same color indicate different quasispecies from the same patient’s sample. Dots on the tree representing samples linked by transmission are enclosed in a grey circle. For patients not linked by transmission (those outside of the grey circle), this testing did not find a degree of genetic relatedness above the transmission criteria threshold.
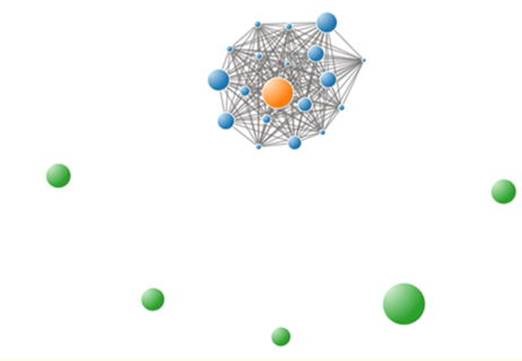
An example GHOST report may be found here: Accurate genetic detection of hepatitis C virus transmissions in outbreak settingsexternal icon
Each patient is represented by a circle of smaller or larger size in proportion with the diversity of virus in that patient’s sample. Pairs or clusters of transmission-linked patient samples are linked by bars.
Read more about GHOST at: HHS Ventures Team Helps Detect Disease Outbreaksexternal icon, GHOST Makes Connections in Hepatitis C Virus Transmission.
References
- Khudyakov, Y., Molecular surveillance of hepatitis C. Antivir Ther, 2012. 17(7 Pt B): p. 1465-70.external icon
- Campo, D.S., et al., Accurate Genetic Detection of Hepatitis C Virus Transmissions in Outbreak Settings. J Infect Dis, 2016. 213(6): p. 957-65. external icon
- Longmire, A.G., et al., GHOST: global hepatitis outbreak and surveillance technology. BMC Genomics, 2017. 18(Suppl 10): p. 916.external icon
- Ramachandran S, Thai H, Forgi JC, et al. A large HCV transmission network enabled a fast-growing HIV outbreak in rural Indiana, 2015. EBioMedicine 2018; 37: 374-81. external icon