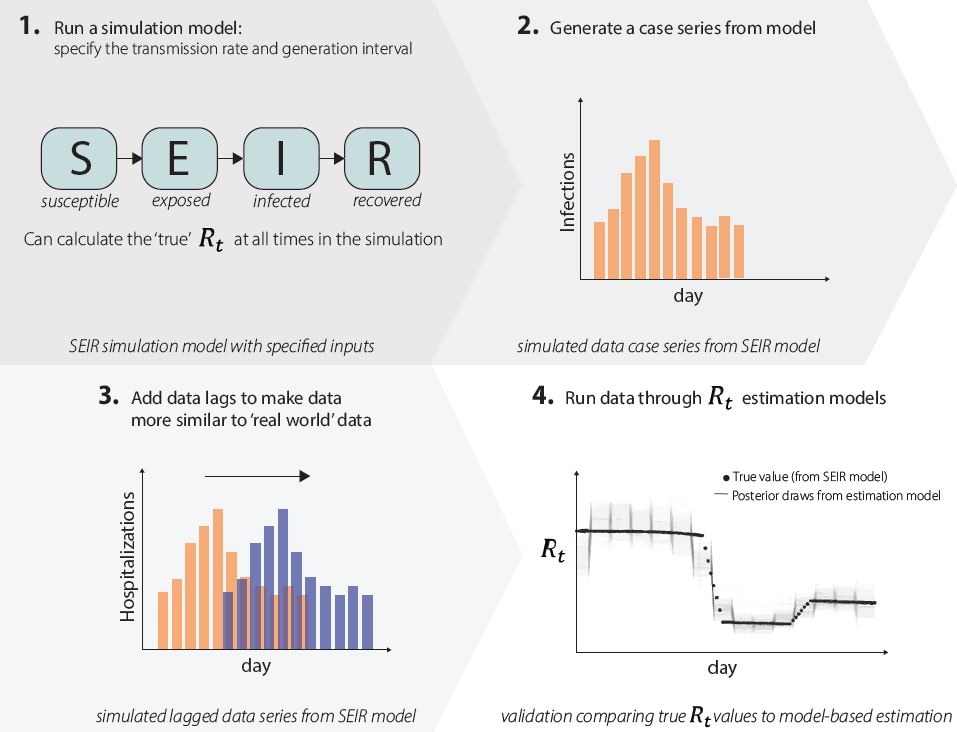
Behind the Model: Improving CDC’s Tools for Assessing Epidemic Growth
At CDC’s Center for Forecasting and Outbreak Analytics, we are building modeling tools and computational pipelines so that we can do complicated data analyses quickly and accurately in response to epidemics. Our goal is to make these tools accessible to federal, state, tribal, territorial, local, and academic partners. One of these efforts is to estimate the time-varying reproductive number, Rt, a measure that helps us quickly assess whether infections are increasing or decreasing.
Measuring transmission with Rt
The basic reproductive number, R0 (pronounced R-naught), is defined as the expected number of new infections caused by each infected person in a fully susceptible population and in the absence of interventions. R0 is an important theoretical concept in epidemiology, but in the real world, a fully susceptible population rarely exists. The time-varying effective reproductive number is known as Rt. When Rt is above one, infections are increasing, and when Rt is below one, infections are decreasing.
- What Rt can tell us: Rt can tell us whether infections are increasing, decreasing, or remaining stable, and is an additional tool to help public health practitioners prepare and respond.
- What Rt cannot tell us: Rt cannot tell us about the underlying burden of disease, just the trend of transmission. An Rt < 1 does not mean that transmission is low, just that infections are decreasing. It is useful to look at respiratory disease activity in conjunction with Rt.
An epidemic’s growth and decline are driven by underlying changes in transmission over time. Early in an epidemic of a novel disease, when everyone is susceptible, transmission rates are usually highest, and then decline as people change their behavior to avoid infection or gain immunity through infection or immunization. The peak of an epidemic is a turning point that occurs when transmission falls below a critical threshold where, on average, each infected person no longer causes more than one new infection.
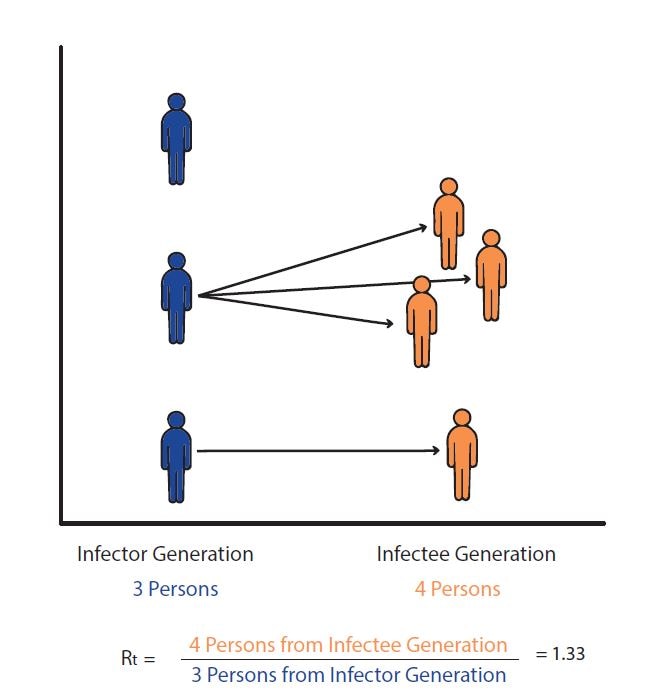
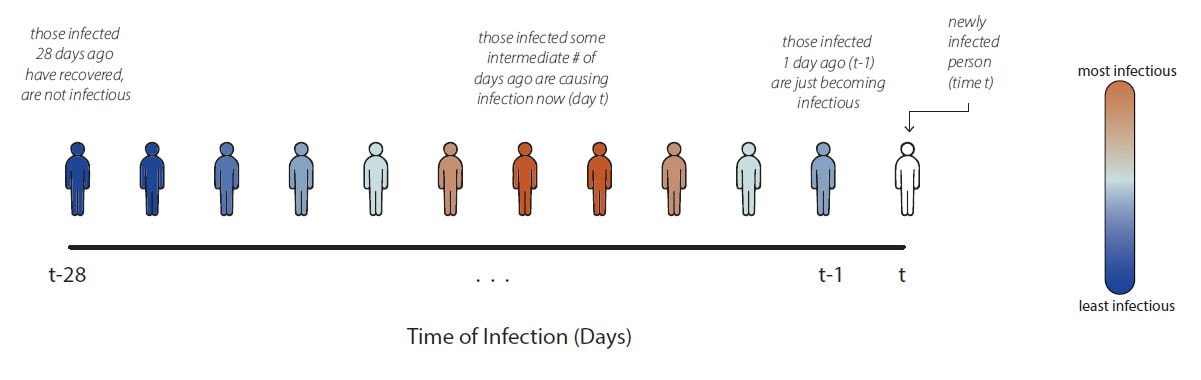
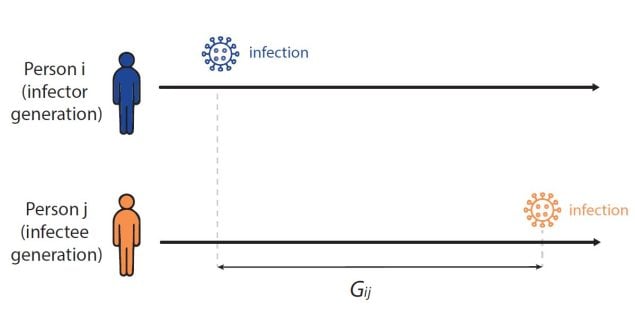
Rt is defined as the average number of new infections caused by each infected person at time t, usually measured in days. Rt tells us whether at that time, infections are increasing, decreasing, or staying relatively flat. We estimate Rt from data, and it accounts for current levels of population susceptibility, interventions, and behavior at the time the underlying infections occurred (Fig. 1).
During an epidemic, Rt estimates provide information about the growth status of the epidemic and can be used to forecast short-term changes in cases, hospitalizations, or deaths, and to assess the effectiveness of interventions designed to slow transmission.
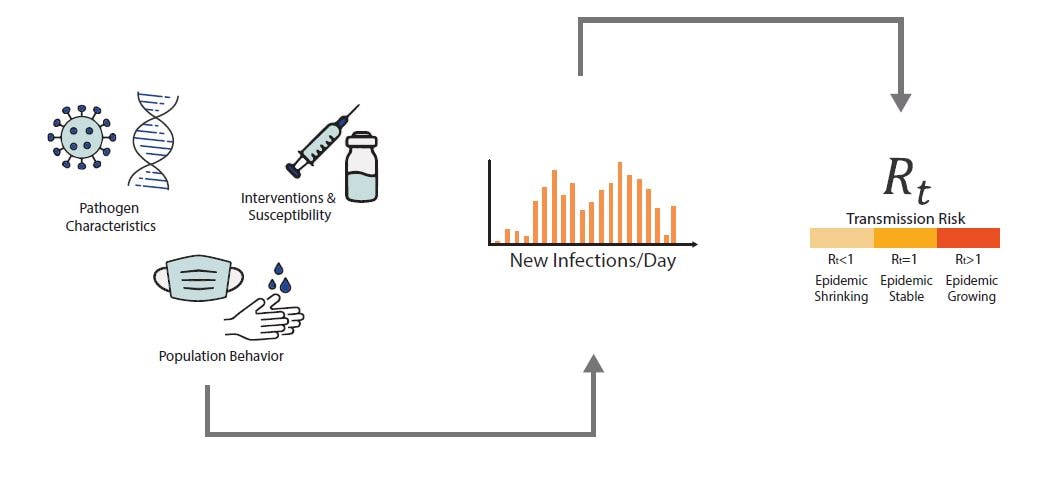
Fig 1. A combination of factors will determine the number of new infections per day (an unobserved quantity) including a pathogen’s characteristics such as infectiousness or severity, public health interventions and susceptibility in the population, and population behaviors and connectivity. Using models combined with observed data, we can determine if infections are increasing, decreasing, or staying relatively flat – as measured by Rt.
Rt is a transmission metric that estimates the ratio of infected to infectors in an epidemic at a particular point in time. Rt estimates help inform situational awareness, giving clues as to how quickly an epidemic is likely to increase or decrease in the near future. Rt estimates can even form the backbone of quantitative short-term epidemic forecasts. To be useful for decision making, Rt estimates need to be accurate, accounting for time lags because transmission events causing cases now occurred days to weeks ago.
Especially in a novel outbreak, it is essential to know whether the epidemic has started, if we are nearing a peak, and/or if transmission has begun to decline. Rt allows policy-makers and public health decision-makers to assess the impact of interventions, because it estimates how transmission rates have changed over time, and to assess the intensity of spread, because it directly reflects growth in infections.
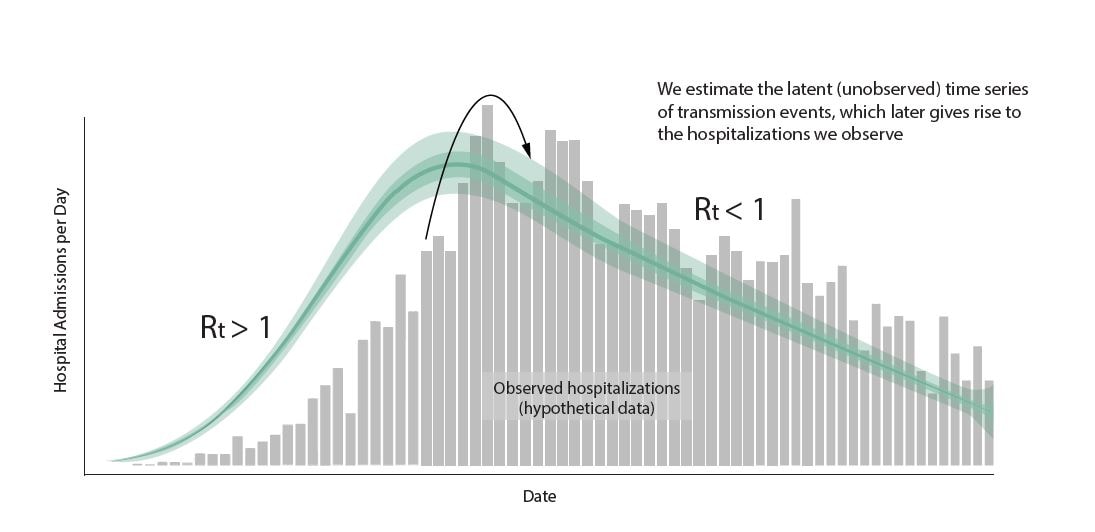
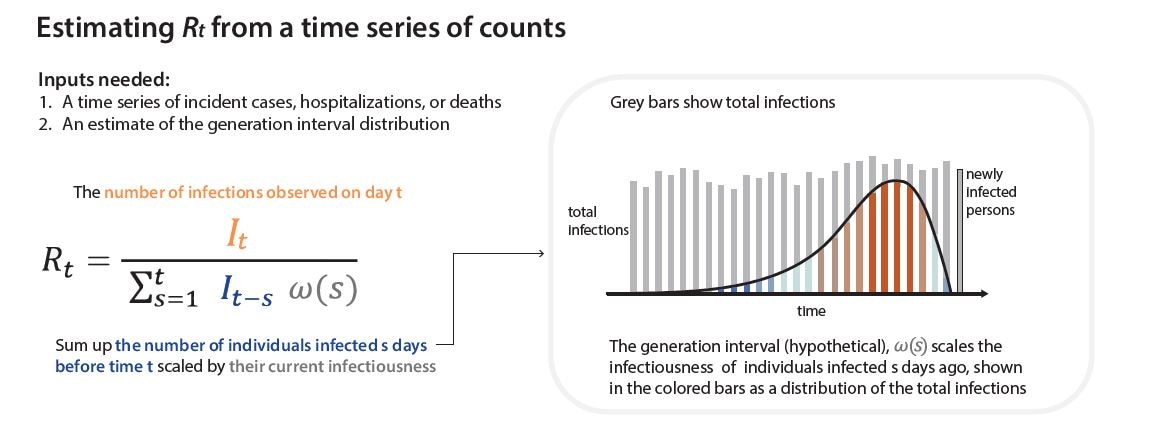
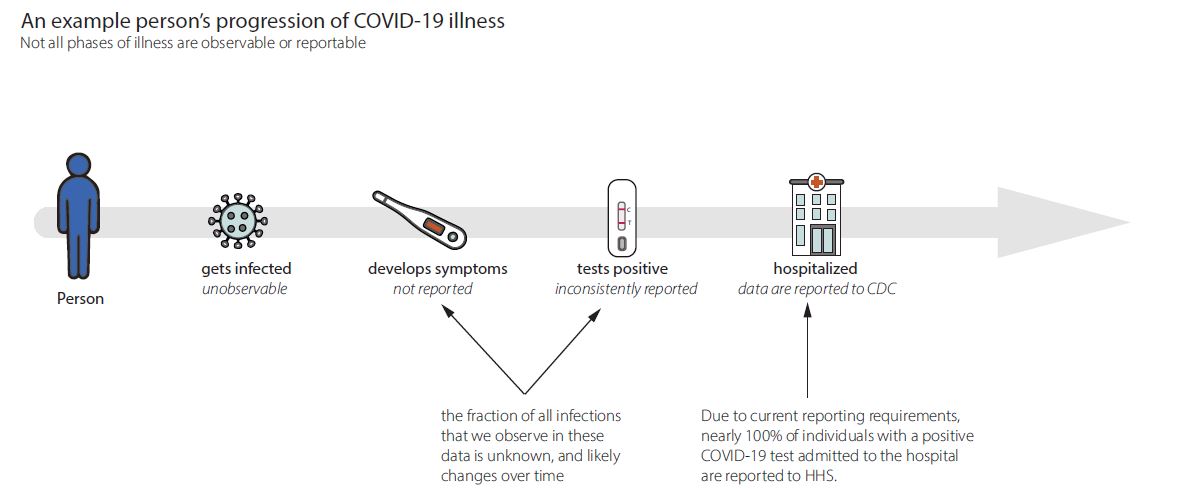
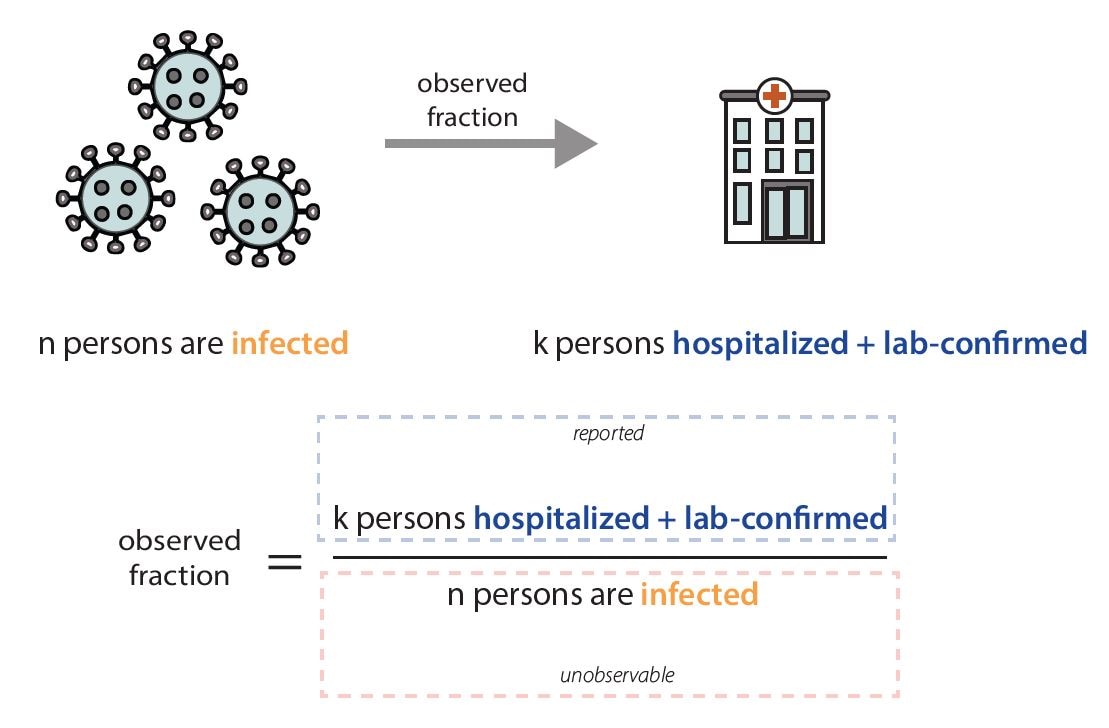
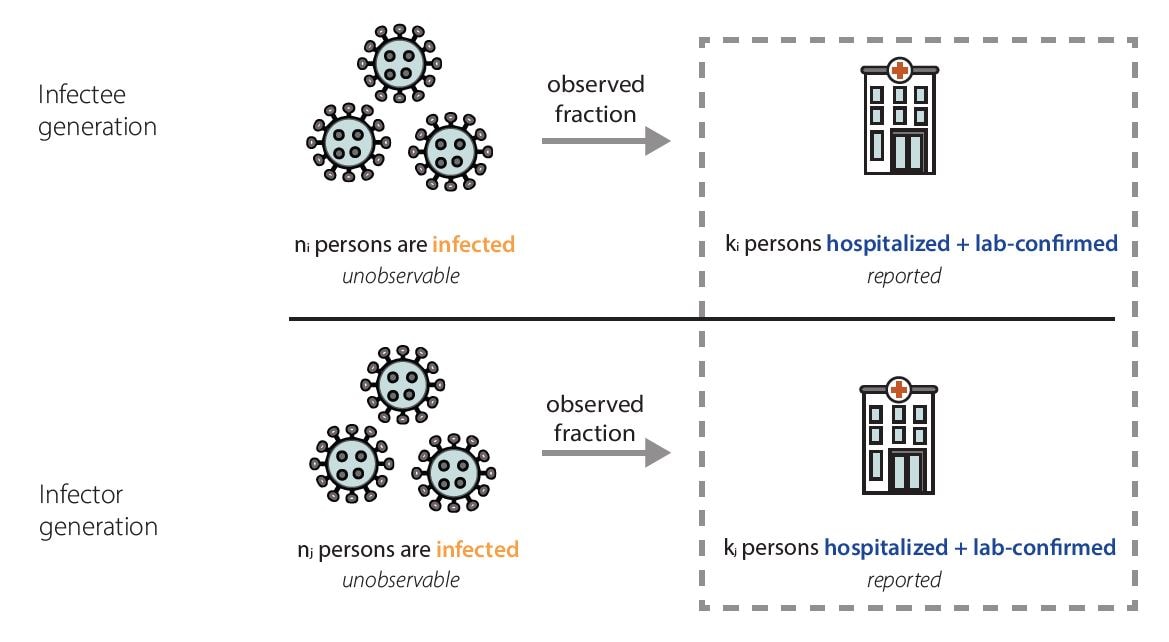
Fig. 10 Shows estimated timeseries of infections of individuals that will go on to be hospitalized, represented in the green line, surrounded by credible interval bands, with observed hospitalizations depicted in grey bars. To estimate Rt, we use a model that adjusts for incomplete observation at the end of the time series, here recent infections have not yet been hospitalized or reported in the data, adjusts for day of week effects, and back-calculates the underlying timing of infections before estimating Rt as the ratio of new infections to infectors. At CDC we use the statistical programming language R to do this data analysis in a single Bayesian model. Graphic modified from EpiNow2 [https://zenodo.org/records/8380568].
What’s next?
We are starting with Rt estimation for respiratory viruses in collaboration with the National Center for Immunization and Respiratory Diseases, but in the long term, we plan to build a well-tested analytic infrastructure that we could use to estimate Rt for a novel pathogen in a future infectious disease epidemic. Even for something like Rt, where the quantity we’re trying to estimate is conceptually relatively simple, it takes incredible care and complicated modeling tools to adjust data as they are observed and obtain accurate estimates quickly. We are also exploring new models that will allow us to combine wastewater data with other signals when we estimate Rt. It is incredibly difficult to build these kinds of analytic pipelines on the fly. Investing the time now to build good infrastructure and think through the problems we can anticipate will leave us better prepared for the next infectious disease epidemic.

I lead the Nowcasting and Natural History Team in the Predict Division. We develop models to help CDC understand what’s going on right now in an epidemic from incomplete or preliminary data in real time. Before joining CDC, I was a PhD student at UCLA, a James S. McDonnell Foundation Complex Systems Fellow, and a research scientist at the University of Chicago.

I am a Data Scientist in the Predict Division on the Nowcasting and Natural History Team where I work on nowcasting and Rt estimation. Previously, I worked for the Rockefeller Foundation and attended Georgetown University.

I am an epidemiologist working with the Inform Division on communicating and translating modeling work for varied audiences and developing interpretable data visualizations for public health communication. Previously, I was an EIS Officer at CDC and have a PhD in infectious disease epidemiology from the University of Michigan.

I’m a Health Scientist on the Applied Research and Modeling Team in the Influenza Division in NCIRD. I primarily work on evaluating and visualizing influenza forecasts for the FluSight forecasting hub, modeling influenza disease outcomes mediated by antivirals and vaccines, and analyzing public health datasets. My previous experience includes infectious and non-infectious disease surveillance.

I’m an infectious disease researcher who specializes in real-time analysis, forecasting, semi-mechanistic modeling, and open-source tool development. I am currently a contractor in the Predict Division on the Nowcasting and Natural History Team. I am also part of the Epiforecasts group at the London School of Hygiene and Tropical Medicine.

I am a data scientist on the Nowcasting and Natural History Team in the Predict Division, focusing on creating models for nowcasting, and data pipelines to support those models. Previously, I worked at the Johns Hopkins Applied Physics Lab doing high fidelity modeling and simulation, data analysis, and data tools development.

I am a Mathematical Statistician leading influenza modeling and forecasting efforts in the Applied Research and Modeling Team in the Influenza Division, such as the FluSight influenza forecasting collaboration. I was a founding member of the COVID-19 Scenario Modeling Hub and have experience modeling infectious disease dynamics in multiple pathogen systems.