Strain of Multidrug-Resistant Salmonella Newport Remains Linked to Travel to Mexico and U.S. Beef Products — United States, 2021–2022
Weekly / November 10, 2023 / 72(45);1225–1229
Laura Ford, PhD1; Zachary Ellison1,2; Colin Schwensohn, MPH1; Isabel Griffin, PhD1,3; Meseret G. Birhane, MPH1; Andrea Cote, DVM4; Gamola Z. Fortenberry, PhD4; Selam Tecle, MPH5; Jeffrey Higa, MPH5; Samantha Spencer, MPH6; Brianna Patton, MPH7; Jaimini Patel, MPH8; Julie Dow, MPH9; Azarnoush Maroufi, MPH10; Amy Robbins, MPH11; Danielle Donovan, MS12; Conor Fitzgerald, MPH13; Sierra Burrell, DVM14; Beth Tolar, MS1; Jason P. Folster, PhD1; Laura A. Cooley, MD1; Louise K. Francois Watkins, MD1 (View author affiliations)
View suggested citationSummary
What is already known about this topic?
CDC monitors illness from a persisting, multidrug-resistant strain of Salmonella Newport linked to travel to Mexico, beef products obtained in the United States, and cheese obtained in Mexico.
What is added by this report?
The number of human infections with this strain doubled in 2021 from the 3-year baseline. Travel to Mexico remained a risk factor. Two multistate outbreaks were investigated; beef products obtained in the United States were the suspected vehicle in one outbreak and the confirmed vehicle in the other.
What are the implications for public health practice?
Safe food and drink consumption practices while traveling and at home, and interventions along the food production chain to ensure beef product safety might help prevent illness.
Altmetric:
Abstract
In 2016, CDC identified a multidrug-resistant (MDR) strain of Salmonella enterica serotype Newport that is now monitored as a persisting strain (REPJJP01). Isolates have been obtained from U.S. residents in all 50 states and the District of Columbia, linked to travel to Mexico, consumption of beef products obtained in the United States, or cheese obtained in Mexico. In 2021, the number of isolates of this strain approximately doubled compared with the 2018–2020 baseline and remained high in 2022. During January 1, 2021– December 31, 2022, a total of 1,308 isolates were obtained from patients, cattle, and sheep; 86% were MDR, most with decreased susceptibility to azithromycin. Approximately one half of patients were Hispanic or Latino; nearly one half reported travel to Mexico during the month preceding illness, and one third were hospitalized. Two multistate outbreak investigations implicated beef products obtained in the United States. This highly resistant strain might spread through travelers, animals, imported foods, domestic foods, or other sources. Isolates from domestic and imported cattle slaughtered in the United States suggests a possible source of contamination. Safe food and drink consumption practices while traveling and interventions across the food production chain to ensure beef safety are necessary in preventing illness.
Introduction
In 2016, CDC identified a multidrug-resistant (MDR) strain of Salmonella enterica serotype Newport that is now monitored as a persisting strain* named REPJJP01 and includes isolates from U.S. residents in all 50 states and the District of Columbia. A 2018–2019 investigation found that persons with these infections reported traveling to Mexico or consuming beef products obtained in the United States or cheese obtained in Mexico (1). During 2021, CDC’s PulseNet database received an increase of REPJJP01 reports, which prompted this investigation. This report analyzed isolates identified during 2021–2022.
Methods
Isolate Sequencing and Determination of Antimicrobial Resistance
Salmonella isolates underwent whole genome sequencing at U.S. public health laboratories and federal agencies, and the resulting sequence data were uploaded to the PulseNet Salmonella National Database and analyzed by core genome multilocus sequence typing (cgMLST) (2). CDC defined REPJJP01 as a strain of S. Newport with a range of 0–21 allele differences by cgMLST. Isolate genomes were screened for resistance determinants and assigned predicted resistance patterns using ResFinder† drug keys as part of surveillance through the National Antimicrobial Resistance Monitoring System. Isolates were defined as MDR if they contained resistance determinants for three or more antimicrobial classes.
Data Analyses
Public health officials obtained travel and food exposure information from interviews and medical records of patients with the REPJJP01 strain isolated; some data were not available for all patients. Characteristics of patients with illness onset dates§ and food and animal samples with collection dates during January 1, 2021–December 31, 2022, were summarized, and proportions were compared using chi-square tests; p<0.05 was considered statistically significant. R statistical software (version 4.2.3; R Foundation) was used to conduct these analyses. CDC investigated multistate outbreaks among nontravelers with support from local and state health departments and the U.S. Department of Agriculture’s Food Safety and Inspection Service. This activity was reviewed by CDC, deemed not research, and was conducted consistent with applicable federal law and CDC policy.¶
Results
Trends in Isolate Detection
In 2021, the total number of human isolates (641) approximately doubled from the average annual baseline number of cases detected during 2018–2020** (315) and remained high in 2022 (641) (Figure). Nonhuman isolates included 25 from cattle (beef products and cecal†† samples) and one from a sheep (cecal sample) from 13 U.S. states.§§
Patient and Isolate Characteristics
The median patient was aged 37.5 years (IQR = 23–55 years), 52% were female, 56% were Hispanic or Latino (Hispanic), and 19% and 17% were California and Texas residents, respectively (Table). Among all patients with hospitalization data available (721), one third (247, 33%) were hospitalized, and two died (<1%). The percentage of Hispanic patients who were hospitalized (40%) was higher than that of non-Hispanic White patients (24%, p<0.001) or non-Hispanic patients of other races (22%, p = 0.03). Most patient isolates (1,141, 89%) were resistant or had decreased susceptibility to at least one antibiotic recommended for empiric or alternative treatment (3–5), and 1,110 (87%) were MDR. Most food and cecal isolates also had antimicrobial resistance; 65% of food and cecal isolates were MDR.
Travel to Mexico remained a risk factor for infection during 2021–2022. Among 721 patients with known travel history, nearly one half (344, 48%) reported traveling to Mexico in the month before illness onset. Compared with persons without a recent history of travel to Mexico, fewer travelers to Mexico were Hispanic (43% versus 64%, p<0.001) or were hospitalized (24% versus 37%, p<0.001). Reported travel to Mexico was more prevalent in 2022 (53%) than in 2021 (39%) (p<0.001). Eleven ill nontravelers reported eating foods, including queso fresco or dried beef, purchased in Mexico by family or friends.
Outbreak Investigations
A multistate 75-patient outbreak was investigated during fall 2021; 80% of patients in that outbreak were Hispanic, and beef, including dried beef, was the suspected vehicle. A second multistate outbreak that included 22 non-Hispanic patients was investigated during fall 2022. A strain of S. Newport indistinguishable from the clinical isolates was isolated in a sample of leftover ground beef from a patient. However, because the beef could not be traced back definitively to a common source, no regulatory action could be taken. Six subclusters of REPJJP01 isolates related within 0–2 allele differences during 2021–2022 contained at least one clinical and one beef isolate; however, these small clusters did not include sufficient cases to conduct an epidemiologic investigation that could implicate a product.
Discussion
During 2021–2022, a total of 1,282 U.S. residents had culture-confirmed infections caused by REPJJP01, an MDR strain of S. Newport. The total number of cases of illness is likely much larger: an additional 29 cases of Salmonella are estimated for each culture-confirmed case (6). Further, the sharp increase in cases of REPJJP01 during 2021 occurred despite an overall decrease in Salmonella reports during the COVID-19 pandemic (7). Patient exposures and outbreak sources resembled those identified in the 2018–2019 investigation (1), including travel to Mexico, consumption of beef and cheese products purchased in Mexico, and consumption of beef products obtained in the United States.
This strain might spread to the United States through returned travelers from Mexico, cattle born or raised in Mexico and slaughtered in the United States, or beef or cheese imported from Mexico. The increase in cases during 2021–2022 appears driven partially by travelers; the 48% of patients who reported travel to Mexico is high compared with the 9% of all patients with nontyphoidal Salmonella reporting any international travel.¶¶ In September 2022, CDC posted a Level 1 Travel Health Notice to alert travelers to Mexico about the risk for infection and to provide advice for prevention.*** Travelers should practice safe eating, drinking, cooking, and food handling habits to reduce their chance of becoming ill. Among nontravelers, the high proportion of Hispanic patients and patient reports of eating beef or cheese purchased by family or friends in Mexico suggests that beef or cheese imported from Mexico might be an important source of illness.†††
The REPJJP01 strain might also spread within the United States through animals or beef products. The strain has been isolated in samples collected in federally regulated slaughter and processing establishments throughout the United States and import establishments on the Southern border. The isolation of this strain from a beef product collected during a 2022 multistate outbreak investigation suggests cattle infected with Salmonella in the United States might serve as a potential source of contamination. During outbreaks, the source of implicated beef products is difficult to identify because patients report consuming beef products both inside and outside the home, shopper history records or receipts are often not available, and traceback is complex, with challenges including incomplete retail grinding logs and multiple products combined in retail products (8). Steps to prevent contamination of beef products might occur across the food production chain, including at preharvest (e.g., herd and biosecurity management), slaughter (e.g., appropriate sanitary dressing procedures and application of effective antimicrobial solutions), processing, and retail (8). Consumers should wash hands and surfaces after preparing raw meat at home and use a thermometer to ensure appropriate cooking temperatures are reached (9).
Some persons infected with REPJJP01 became seriously ill: 33% of all patients, 37% of non-travelers, and 40% of Hispanic patients were hospitalized, compared with 27% of all patients with nontyphoidal Salmonella infections reporting hospitalization in FoodNet data during 2021–2022. The high hospitalization rate is consistent with studies indicating that patients with antimicrobial-resistant Salmonella infections are more likely to be hospitalized (10). The increase in infections from this MDR strain is concerning because it limits treatment options, has more severe outcomes, and creates opportunities for resistance genes to spread. Clinicians should be aware of the potential for multidrug resistance in recent travelers to Mexico with salmonellosis and should order susceptibility testing to guide antimicrobial selection when treatment is warranted.§§§
Limitations
The findings in this report are subject to at least three limitations. First, the number of illnesses recorded by the public health system is an underestimate of the total number of illnesses. Second, not all patients were able to be interviewed, limiting the ability to collect exposure information. Finally, the proportion of patients reporting travel might be an underestimate, because some patients were only asked about travel during the 4 or 7 days before illness onset.
Implications for Public Health Practice
CDC continues to work with local and state health departments and federal partners to investigate cases of this strain to identify sources of infection and prevent illness. Interventions along the food production chain to ensure beef safety (8) and prevention measures among producers, consumers, and travelers might help to reduce illness from this persisting MDR strain. Consumers should eat beef only if it is cooked thoroughly. Travelers to Mexico should follow food safety practices while abroad (https://wwwnc.cdc.gov/travel/page/food-water-safety); to reduce the chances of illness, travelers should avoid beef (including beef jerky or dried beef) and other foods that are prepared or sold by street vendors. Further research is needed to understand the consequences of decreased susceptibility to azithromycin on clinical outcomes for patients receiving this agent.
Acknowledgments
Epidemiologist and laboratory partners in state and local health and agriculture departments.
Corresponding author: Laura Ford, qdz4@cdc.gov.
1Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, CDC; 2ASRT, Inc., Smyrna, Georgia; 3Epidemic Intelligence Service, CDC; 4Food Safety and Inspection Service, U.S. Department of Agriculture, Washington, DC; 5California Department of Public Health; 6Texas Department of State Health Services; 7Pueblo Department of Public Health and Environment, Pueblo, Colorado; 8Los Angeles County Department of Public Health, Los Angeles, California; 9Illinois Department of Public Health; 10Health & Human Services Agency, San Diego, California; 11New York State Department of Health; 12Michigan Department of Health & Human Services; 13Arizona Department of Health Services; 14Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Washington, DC.
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
* This strain is monitored as a reoccurring, emerging, or persisting (REP) strain; “persisting” is defined as causing illness consistently during a long period. https://www.cdc.gov/ncezid/dfwed/outbreak-response/rep-strains.html
† https://github.com/tseemann/shovill; https://github.com/phac-nml/staramr; https://github.com/StaPH-B/resistanceDetectionCDC
§ If illness onset date was unknown, it was presumptively calculated as the isolation date minus 3 days.
¶ 45 C.F.R. part 46, 21 C.F.R. part 56; 42 U.S.C. Sect. 241(d); 5 U.S.C. Sect. 552a; 44 U.S.C. Sect.3501 et seq.
** The 3-year baseline is the average number of culture-confirmed isolates during 2018–2020.
§§ No samples were imported during 2021–2022.
¶¶ https://wwwn.cdc.gov/foodnetfast
*** https://wwwnc.cdc.gov/travel/notices/watch/salmonella-newport-mexico
††† https://ask.usda.gov/s/article/Products-for-personal-consumption
§§§ https://www.cdc.gov/salmonella/general/technical/newport.html
References
- Plumb ID, Schwensohn CA, Gieraltowski L, et al. Outbreak of Salmonella Newport infections with decreased susceptibility to azithromycin linked to beef obtained in the United States and soft cheese obtained in Mexico—United States, 2018–2019. MMWR Morb Mortal Wkly Rep 2019;68:713–7. https://doi.org/10.15585/mmwr.mm6833a1 PMID:31437141
- Leeper MM, Tolar BM, Griswold T, et al. Evaluation of whole and core genome multilocus sequence typing allele schemes for Salmonella enterica outbreak detection in a national surveillance network, PulseNet USA. Front Microbiol 2023;14:1254777. https://doi.org/10.3389/fmicb.2023.1254777 PMID:3708298
- McDermott PF, Tyson GH, Kabera C, et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 2016;60:5515–20. https://doi.org/10.1128/AAC.01030-16 PMID:27381390
- Shane AL, Mody RK, Crump JA, et al. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 2017;65:e45–80. https://doi.org/10.1093/cid/cix669 PMID:29053792
- Meltzer PS, Kallioniemi A, Trent JM. Chromosome alterations in human solid tumors [Chapter 9]. In: Vogelstein B, Kinzler KW, eds. The genetic basis of human cancer. New York, NY: McGraw-Hill; 2002:93–113.
- Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 2011;17:7–15. https://doi.org/10.3201/eid1701.P11101 PMID:21192848
- Collins JP, Shah HJ, Weller DL, et al. Preliminary incidence and trends of infections caused by pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2016–2021. MMWR Morb Mortal Wkly Rep 2022;71:1260–4. https://doi.org/10.15585/mmwr.mm7140a2 PMID:36201372
- Canning M, Birhane MG, Dewey-Mattia D, et al. Salmonella outbreaks linked to beef, United States, 2012–2019. J Food Prot 2023;86:100071. https://doi.org/10.1016/j.jfp.2023.100071 PMID:37028195
- US Department of Health and Human Services. Keep food safe: food safety by type of food. Washington DC: US Department of Health and Human Services; 2019. https://www.foodsafety.gov/keep-food-safe/food-safety-by-type-food#meat
- Varma JK, Molbak K, Barrett TJ, et al. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis 2005;191:554–61. https://doi.org/10.1086/427263 PMID:15655779
 FIGURE. Month and year of illness onset* or sample collection† and source type for isolates of multidrug resistant Salmonella Newport strain REPJJP01 — United States, 2016–2022
FIGURE. Month and year of illness onset* or sample collection† and source type for isolates of multidrug resistant Salmonella Newport strain REPJJP01 — United States, 2016–2022
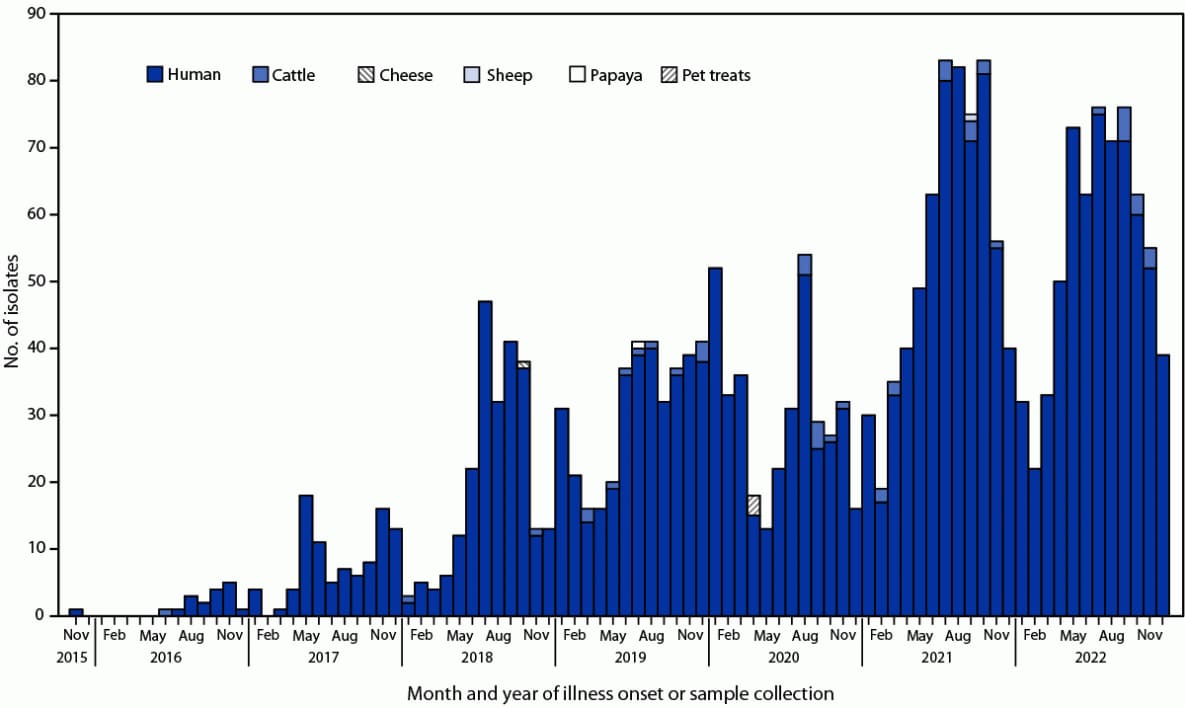
* Some illness onset dates for human cases have been estimated based on the isolation date.
† Nonhuman isolates.
* Denominators may not equal totals for each year due to missing data.
† Persons of Hispanic or Latino (Hispanic) origin might be of any race but are categorized as Hispanic; all racial groups are non-Hispanic.
§ Other source sites include abscess, fluid, iliac aneurysm, rectal swab, synovial fluid, tissue, or wound.
¶ Resistance was determined based on the results of antimicrobial susceptibility testing when available (37); otherwise, resistance was predicted based on whole genome sequencing. Isolates were considered to be resistant to an antimicrobial if the presence of at least one resistance mechanism known to confer resistance to that antimicrobial was present in the isolate’s genome. One isolate in 2021 was not tested by antimicrobial susceptibility testing or analyzed for resistance determinants and is not included in the denominator.
** Interpretive criteria for azithromycin resistance have not been established for Salmonella serotypes other than serotype Typhi. Therefore, the presence of resistance determinants for azithromycin should not be used to predict clinical efficacy.
†† Includes isolates with interpretation of “intermediate” or isolates carrying a single quinolone resistance gene (878; 89%); a single gene might result in interpretation of “intermediate” for ciprofloxacin on antimicrobial susceptibility testing.
Suggested citation for this article: Ford L, Ellison Z, Schwensohn C, et al. Strain of Multidrug-Resistant Salmonella Newport Remains Linked to Travel to Mexico and U.S. Beef Products — United States, 2021–2022. MMWR Morb Mortal Wkly Rep 2023;72:1225–1229. DOI: http://dx.doi.org/10.15585/mmwr.mm7245a3.
MMWR and Morbidity and Mortality Weekly Report are service marks of the U.S. Department of Health and Human Services.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of
Health and Human Services.
References to non-CDC sites on the Internet are
provided as a service to MMWR readers and do not constitute or imply
endorsement of these organizations or their programs by CDC or the U.S.
Department of Health and Human Services. CDC is not responsible for the content
of pages found at these sites. URL addresses listed in MMWR were current as of
the date of publication.
All HTML versions of MMWR articles are generated from final proofs through an automated process. This conversion might result in character translation or format errors in the HTML version. Users are referred to the electronic PDF version (https://www.cdc.gov/mmwr) and/or the original MMWR paper copy for printable versions of official text, figures, and tables.
Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.