Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2015–2018
Weekly / April 26, 2019 / 68(16);369–373
Please note: An erratum has been published for this report. To view the erratum, please click here.
Danielle M. Tack, DVM1; Ellyn P. Marder, MPH1; Patricia M. Griffin, MD1; Paul R. Cieslak, MD2; John Dunn, DVM3; Sharon Hurd, MPH4; Elaine Scallan, PhD5; Sarah Lathrop, PhD6; Alison Muse, MPH7; Patricia Ryan, MD8; Kirk Smith, DVM9; Melissa Tobin-D’Angelo, MD10; Duc J. Vugia, MD11; Kristin G. Holt, DVM12; Beverly J. Wolpert, PhD13; Robert Tauxe, MD1; Aimee L. Geissler, PhD1 (View author affiliations)
View suggested citationSummary
What is already known about this topic?
The incidence of foodborne infections has remained largely unchanged. Clinical laboratories are increasingly using culture-independent diagnostic tests (CIDTs) to detect enteric infections. CIDTs benefit public health surveillance by identifying pathogens not routinely detected by previous methods but complicate data interpretation.
What is added by this report?
The incidence of most infections increased during 2018 compared with 2015–2017; this might be partially attributable to increased CIDT use. The incidence of Cyclospora infections increased markedly, in part related to large outbreaks associated with produce. The number of human infections caused by Campylobacter and Salmonella, especially serotype Enteritidis, remains high.
What are the implications for public health practice?
As use of CIDTs increases, it is important to obtain and subtype isolates and interview ill persons to monitor prevention efforts and develop more targeted prevention and control measures to make food safer and decrease human illness.
Altmetric:
Foodborne diseases represent a major health problem in the United States. The Foodborne Diseases Active Surveillance Network (FoodNet) of CDC’s Emerging Infections Program monitors cases of laboratory-diagnosed infection caused by eight pathogens transmitted commonly through food in 10 U.S. sites.* This report summarizes preliminary 2018 data and changes since 2015. During 2018, FoodNet identified 25,606 infections, 5,893 hospitalizations, and 120 deaths. The incidence of most infections is increasing, including those caused by Campylobacter and Salmonella, which might be partially attributable to the increased use of culture-independent diagnostic tests (CIDTs). The incidence of Cyclospora infections increased markedly compared with 2015–2017, in part related to large outbreaks associated with produce (1). More targeted prevention measures are needed on produce farms, food animal farms, and in meat and poultry processing establishments to make food safer and decrease human illness.
FoodNet conducts active, population-based surveillance for laboratory-diagnosed infections caused by Campylobacter, Cyclospora, Listeria, Salmonella, Shiga toxin–producing Escherichia coli (STEC), Shigella, Vibrio, and Yersinia in 10 sites covering 15% of the U.S. population (approximately 49 million persons in 2017). FoodNet is a collaboration among CDC, 10 state health departments, the U.S. Department of Agriculture’s Food Safety and Inspection Service (USDA-FSIS), and the Food and Drug Administration (FDA). Bacterial infections are defined as isolation of the bacterium from a clinical specimen or detection of pathogen antigen, nucleic acid sequences, or, for STEC,† Shiga toxin or Shiga toxin genes. Listeria cases are defined as isolation of L. monocytogenes or detection of its nucleic acid sequences from a normally sterile site or from placental or fetal tissue in cases of miscarriage or stillbirth. Cyclospora infections are defined as detection of the parasite from a clinical specimen by direct fluorescent antibody, polymerase chain reaction, or light microscopy. Hospitalizations occurring within 7 days of specimen collection are attributed to the infection, as is the patient’s vital status at hospital discharge, or 7 days after specimen collection if the patient was not hospitalized.
Incidence per 100,000 population was calculated by dividing the number of infections in 2018 by U.S. Census estimates of the surveillance area population for 2017. A negative binomial model with 95% confidence intervals (CIs) was calculated using SAS (version 9.4; SAS Institute) to estimate changes in incidence.
Surveillance for physician-diagnosed postdiarrheal hemolytic uremic syndrome, a complication of STEC infection characterized by renal failure, thrombocytopenia, and microangiopathic anemia, is conducted through a network of nephrologists and infection preventionists and by hospital discharge data review. This report includes pediatric hemolytic uremic syndrome cases (those occurring in persons aged <18 years) identified during 2017, the most recent year for which data are available.
Cases of Infection, Incidence, and Trends
During 2018, FoodNet identified 25,606 cases of infection, 5,893 hospitalizations, and 120 deaths. The incidence of infection (per 100,000 population) was highest for Campylobacter (19.5) and Salmonella (18.3), followed by STEC (5.9), Shigella (4.9), Vibrio (1.1), Yersinia (0.9), Cyclospora (0.7), and Listeria (0.3) (Table). Compared with 2015–2017, the incidence significantly increased for Cyclospora (399%), Vibrio (109%), Yersinia (58%), STEC (26%), Campylobacter (12%), and Salmonella (9%). The number of bacterial infections diagnosed by CIDT (with or without reflex culture§) increased 65% in 2018 compared with the average annual number diagnosed during 2015–2017; the increase ranged from 29% for STEC to 311% for Vibrio (Figure 1). In 2018, the percentage of infections diagnosed by DNA-based syndrome panels was highest for Yersinia (68%) and Cyclospora (67%), followed by STEC (55%), Vibrio (53%), Shigella (48%), Campylobacter (43%), Salmonella (33%), and was lowest for Listeria (2%). In 2018, a reflex culture was attempted on 75% of specimens with positive CIDT results, ranging from 64% for Campylobacter to 100% for Listeria (Figure 1). The percentage of specimens with a reflex culture in 2018 was 14% higher than that during 2015–2017, ranging from a 7% decrease for STEC to a 55% increase for Shigella (Figure 2). Among specimens with reflex culture in 2018, the percentage that yielded the pathogen was highest for Listeria (100%) and Salmonella (86%), followed by STEC (64%), Campylobacter (59%), Shigella (56%), Yersinia (50%), and Vibrio (37%) (Figure 1) (Figure 2).
Among 7,013 (87%) serotyped Salmonella isolates, the three most common were Enteritidis (2.6 per 100,000 population), Newport (1.6), and Typhimurium (1.5), similar to those during 2015–2017. Among 1,570 STEC isolates tested, 440 (28%) were determined to be O157. Among 662 non-O157 STEC isolates serogrouped, the most common were O103 (31%), O26 (28%), and O111 (24%). The incidence compared with 2015–2017 remained unchanged for both O157 and non-O157 STEC.
FoodNet identified 54 cases of postdiarrheal hemolytic uremic syndrome in children (0.49 cases per 100,000) during 2017; 36 (67%) occurred among children aged <5 years (1.22 cases per 100,000). Incidence was not significantly different compared with that during 2014–2016.
Discussion
Campylobacter has been the most commonly identified infection in FoodNet since 2013. It causes diarrhea, sometimes bloody, and 18% of persons are hospitalized. A rare outcome of Campylobacter infection is Guillain-Barré syndrome, a type of autoimmune-mediated paralysis. Poultry is a major source of Campylobacter (2). In August 2018, FSIS began using a new testing method; in a study of that method, Campylobacter was isolated from 18% of chicken carcasses and 16% of chicken parts sampled (3). FSIS currently makes aggregated test results available and intends to update performance standards for Campylobacter contamination.
The incidence of infections with Enteritidis, the most common Salmonella serotype, has not declined in over 10 years. Enteritidis is adapted to live in poultry, and eggs are an important source of infection (4). By 2012, FDA had implemented the Egg Safety Rule,¶ which requires preventive measures during the production of eggs in poultry houses and requires subsequent refrigeration during storage and transportation, for all farms with ≥3,000 hens. In 2018, a multistate outbreak of Enteritidis infections was traced to eggs from a farm that had not implemented the required egg safety measures after its size reached ≥3,000 hens (5). Chicken meat is also an important source of Enteritidis infections (4). In December 2018, FSIS reported that 22% of establishments that produce chicken parts failed to meet the Salmonella performance standard (USDA-FSIS Salmonella verification testing program**). The percentage of samples of chicken meat and intestinal contents that yielded Enteritidis were similar in 2018 to those during 2015–2017 (USDA-FSIS, unpublished data). In contrast, a decline in serotype Typhimurium isolated from the same sources was observed during the same period. This trend coincides with declines in Typhimurium human illnesses. Changes in poultry production practices, including vaccination against Typhimurium, might have resulted in these declines (6). In the United Kingdom, vaccination of both broiler and layer chickens against Enteritidis, along with improved hygiene, was followed by a marked decrease in human Enteritidis infections (7).
Produce is a major source of foodborne illnesses (2). During 2018, romaine lettuce was linked to two multistate outbreaks of STEC O157 infections (8). The marked increase in reported Cyclospora infections was likely attributable to several factors including produce outbreaks and continued adoption of DNA-based syndrome panel tests (1). Improved agricultural practices are needed to prevent produce-associated infections. FDA provides technical assistance to task forces created by the produce industry, to determine how to prevent contamination of romaine lettuce and facilitate outbreak investigations by improving product labeling and traceability. In 2018, FDA expanded surveillance sampling of foreign and domestically grown produce to assess its safety (9). FDA is implementing the Produce Safety Rule,†† with routine inspections of large produce farms planned this spring. Because produce is a major component of a healthy diet and is often consumed raw, making it safer is important for improving human health (10).
The findings in this report are subject to at least three limitations. First, the changing diagnostic landscape makes interpretation of incidence and trends more complex. Increases in reported incidence might be attributable entirely, or in part, to changes in clinician ordering practices, increased use of DNA-based syndrome panels that identify pathogens not routinely captured by traditional methods, and changes in laboratory practices in response to the availability of these panels. Second, some CIDT results might be false positives. Finally, year-to-year variations, attributable in part to large outbreaks, might not indicate sustained trends.
The need to obtain and subtype isolates from ill persons is becoming an increasing burden to state health departments but is critical for maintaining surveillance to detect and investigate outbreaks, evaluating prevention efforts, and developing targeted control measures. Measures that might decrease foodborne illnesses include enhanced efforts targeting Campylobacter contamination of chicken; strengthening prevention measures during egg production, especially within small flocks; vaccinating poultry against Salmonella serotype Enteritidis; decreasing Salmonella contamination of produce, poultry, and meat; and continued implementation of the Food Safety Modernization Act, specifically FDA’s Produce Safety Rule. FoodNet continues to collect data and develop analytic tools to adjust for changes in diagnostic testing practices and test characteristics. These actions, along with FoodNet’s robust surveillance, provide data to help evaluate the effectiveness of prevention efforts and determine when additional measures are needed.
Acknowledgments
Work group members, Foodborne Diseases Active Surveillance Network (FoodNet), Emerging Infections Program, CDC; Brittany Behm, Robert Breazu, Staci Dixon, Elizabeth Greene, Logan Ray, Hazel Shah, Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, CDC.
Corresponding author: Danielle Tack, dot7@cdc.gov, 404-718-3254.
1Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, CDC; 2Oregon Health Authority; 3Tennessee Department of Health; 4Connecticut Department of Public Health; 5University of Colorado, Boulder, Colorado; 6University of New Mexico, Albuquerque, New Mexico; 7New York State Department of Health; 8Maryland Department of Health; 9Minnesota Department of Health; 10Georgia Department of Public Health; 11California Department of Public Health; 12Food Safety and Inspection Service, U.S. Department of Agriculture, Atlanta, Georgia; 13Center for Food Safety and Applied Nutrition, Food and Drug Administration, Silver Spring, Maryland.
All authors have completed and submitted the ICMJE form for disclosure of potential conflicts of interest. No potential conflicts of interest were disclosed.
* Connecticut, Georgia, Maryland, Minnesota, New Mexico, Oregon, Tennessee, and selected counties in California, Colorado, and New York (https://www.cdc.gov/foodnet).
† STEC cases are defined as identification of Shiga toxin or its genes by any laboratory; it is not possible to distinguish among serogroups using CIDTs.
§ Culture of a specimen with a positive CIDT result.
†† https://www.fda.gov/Food/GuidanceRegulation/FSMA/ucm334114.htm.
References
- Casillas SM, Bennett C, Straily A. Notes from the field: multiple cyclosporiasis outbreaks—United States, 2018. MMWR Morb Mortal Wkly Rep 2018;67:1101–2. CrossRef PubMed
- Interagency Food Safety Analytics Collaboration. Foodborne illness source attribution estimates for 2016 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. Atlanta, GA: US Department of Health and Human Services, CDC; 2018. https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2016-report-TriAgency-508.pdf
- US Department of Agriculture. Constituent update special alert. August 27, 2018. FSIS to implement enrichment method to detect Campylobacter in all raw poultry samples. Washington, DC: US Department of Agriculture; 2018. https://www.fsis.usda.gov/wps/portal/fsis/newsroom/meetings/newsletters/constituent-updates/archive/2018/ConstUpdate082718. Accessed 19 February 2019.
- Gu W, Vieira AR, Hoekstra RM, Griffin PM, Cole D. Use of random forest to estimate population attributable fractions from a case-control study of Salmonella enterica serotype Enteritidis infections. Epidemiol Infect 2015;143:2786–94. CrossRef PubMed
- Ingram A, Ripley D. Multistate outbreak of Salmonella Enteritidis associated with shell eggs. Presented at the 2019 PulseNet/OutbreakNet East Coast Regional Meeting, Tampa, FL; January 15–17, 2019.
- Dórea FC, Cole DJ, Hofacre C, et al. Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Appl Environ Microbiol 2010;76:7820–5. CrossRef PubMed
- O’Brien SJ. The “decline and fall” of nontyphoidal Salmonella in the United Kingdom. Clin Infect Dis 2013;56:705–10. CrossRef PubMed
- CDC. Reports of E. coli outbreak investigations from 2018. Atlanta, GA: US Department of Health and Human Services, CDC; 2018. https://www.cdc.gov/ecoli/2018-outbreaks.html
- Food and Drug Administration. FDA sampling assignment update identifies Cyclospora in herbs. Silver Spring, MD; US Department of Health and Human Services, Food and Drug Administration; 2018. https://www.fda.gov/Food/NewsEvents/ConstituentUpdates/ucm618781.htm
- Willett W, Rockström J, Loken B, et al. Food in the Anthropocene: the EAT-Lancet Commission on healthy diets from sustainable food systems. Lancet 2019;393:447–92. CrossRef PubMed
Abbreviation: CI = confidence interval; IR = incidence rate.
* Connecticut, Georgia, Maryland, Minnesota, New Mexico, Oregon, Tennessee, and selected counties in California, Colorado, and New York.
† Data are preliminary.
§ Per 100,000 population.
¶ Increase or decrease.
** All serogroups were combined because it is not possible to distinguish among them using culture-independent diagnostic tests.
 FIGURE 1. Number of infections diagnosed by culture or culture-independent diagnostic tests (CIDTs), by pathogen, year, and culture status — CDC’s Foodborne Diseases Active Surveillance Network,* 2015–2018†
FIGURE 1. Number of infections diagnosed by culture or culture-independent diagnostic tests (CIDTs), by pathogen, year, and culture status — CDC’s Foodborne Diseases Active Surveillance Network,* 2015–2018†

Abbreviation: STEC = Shiga toxin–producing Escherichia coli.
* Connecticut, Georgia, Maryland, Minnesota, New Mexico, Oregon, Tennessee, and selected counties in California, Colorado, and New York.
† Data for 2018 are preliminary.
 FIGURE 2. Percentage of infections diagnosed by culture-independent diagnostic tests (CIDTs), positive CIDTs with a reflex culture,* and reflex cultures that yielded the pathogen, by pathogen — CDC’s Foodborne Diseases Active Surveillance Network,† 2015–2017 and 2018§
FIGURE 2. Percentage of infections diagnosed by culture-independent diagnostic tests (CIDTs), positive CIDTs with a reflex culture,* and reflex cultures that yielded the pathogen, by pathogen — CDC’s Foodborne Diseases Active Surveillance Network,† 2015–2017 and 2018§
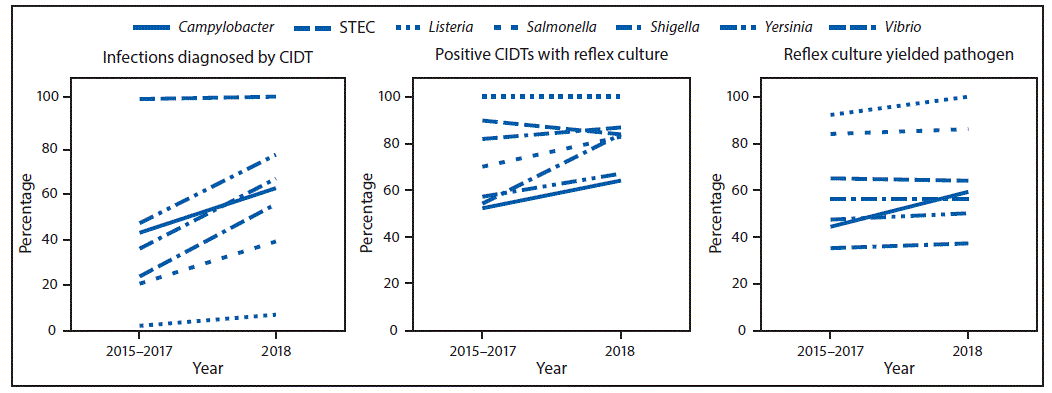
Abbreviation: STEC = Shiga toxin–producing Escherichia coli.
* Culture of a specimen with a positive CIDT result.
† Connecticut, Georgia, Maryland, Minnesota, New Mexico, Oregon, Tennessee, and selected counties in California, Colorado, and New York.
§ Data for 2018 are preliminary.
Suggested citation for this article: Tack DM, Marder EP, Griffin PM, et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2015–2018. MMWR Morb Mortal Wkly Rep 2019;68:369–373. DOI: http://dx.doi.org/10.15585/mmwr.mm6816a2.
MMWR and Morbidity and Mortality Weekly Report are service marks of the U.S. Department of Health and Human Services.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of
Health and Human Services.
References to non-CDC sites on the Internet are
provided as a service to MMWR readers and do not constitute or imply
endorsement of these organizations or their programs by CDC or the U.S.
Department of Health and Human Services. CDC is not responsible for the content
of pages found at these sites. URL addresses listed in MMWR were current as of
the date of publication.
All HTML versions of MMWR articles are generated from final proofs through an automated process. This conversion might result in character translation or format errors in the HTML version. Users are referred to the electronic PDF version (https://www.cdc.gov/mmwr) and/or the original MMWR paper copy for printable versions of official text, figures, and tables.
Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.