MDDR User Guide
User Guide: Molecular Detection of Drug Resistance (MDDR)
in Mycobacterium tuberculosis Complex by DNA Sequencing (Version 3.0)
February 2023
Centers for Disease Control and Prevention Division of Tuberculosis Elimination, Laboratory Branch
1600 Clifton Road, NE Mailstop H17-4
Atlanta, Georgia 30329
e-mail: TBLab@cdc.gov
Telephone: 404-639-2455
Fax: 404-639-5491
Molecular Detection of Drug Resistance (MDDR) can provide rapid and accurate results for detection of drug resistance in Mycobacterium tuberculosis complex (MTBC). This information is critical for the effective treatment of patients suffering from tuberculosis (TB) and initiation of public health interventions. Efforts to treat patients and control the spread of TB can be hindered by the emergence of MTBC resistant to anti-tuberculosis drugs. Additionally, the slow growth rate of MTBC and inherent difficulties associated with some growth-based drug susceptibility testing (DST) methods can serve as impediments to obtaining timely results.
Since September 2009, CDC’s TB laboratory has offered MDDR as a free molecular testing service to all 50 states, U.S. territories, and U.S. Affiliated Pacific Islands using DNA sequencing (pyrosequencing and Sanger sequencing) for the detection of drug-resistance–associated mutations in MTBC (1). In February 2023, the MDDR service will transition to use of primarily targeted next generation sequencing (tNGS) for enhanced early detection of mutations associated with drug resistance. The shift to the tNGS assay allows for deep sequencing of specific (i.e., targeted) genetic loci known to be associated with drug resistance. The methodology is different, but the approach is similar to earlier iterations of MDDR. The change allows for more genetic loci to be examined per test run and better detection of heteroresistance (i.e., mixture of resistant and susceptible populations) that improves the limit of detection (LOD) for resistance-associated mutations. The MDDR tNGS assay was operationalized following a comprehensive evaluation of the tNGS amplicons included in the assay and an associated bioinformatics pipeline. The testing performed is CLIA compliant (https://www.cdc.gov/ncezid/pdf/certificates/infectious-diseases-laboratory-other-atlanta-ga-print-only-version.pdf). Validation included determination of the LOD for heteroresistant populations, intra-assay precision and reproducibility, and clinical sensitivity and specificity. The tNGS assay examines 24 amplicons across 16 genes providing information on more than 12 antituberculosis drugs.
With the implementation of the tNGS assay as part of the MDDR service, CDC will transition from the Escherichia coli to the M. tuberculosis numbering system for reporting of mutations in the rpoB gene. For example, when using the E. coli numbering system, the most commonly reported rpoB mutation associated with rifampin (RIF) resistance is the Ser531Leu mutation. This mutation will now be reported as the Ser450Leu mutation based on the M. tuberculosis numbering system. A graphic developed by the Association of Public Health Laboratories (APHL) is included below to assist with understanding the M. tuberculosis numbering in rpoB for the codons most commonly reported for rifampin resistance (2).
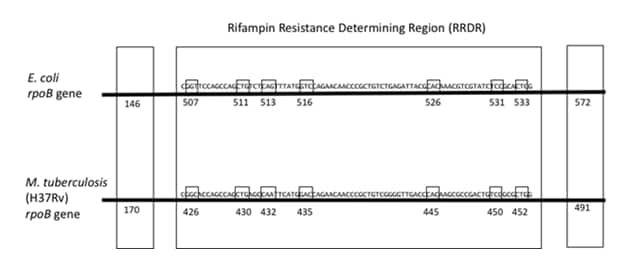
Figure 1. The seven codons in the rifampin resistance determining region (RRDR) frequently associated with RIF resistance are shown for both E. coli and M. tuberculosis. Additionally, the two most common mutations found outside of the RRDR are shown for both species. The M. tuberculosis numbering is minus 81 codons from the E. coli numbering except for the 146/170 codon. Figure is adapted from Andre, 2017 et al and kindly provided by the Association of Public Health Laboratories with permission for use (2,3).
Drug resistance in MTBC is explained by the presence of mutations in specific genes. These mutations often consist of only a single nucleotide change in the DNA sequence (i.e., point mutation). For example, >95% of clinical isolates that are resistant to RIF have a single point mutation in an 81-bp region of the rpoB gene known as the RIF resistance determining region (RRDR) (4). Mutations in this region affect the protein structure of the target so that RIF cannot bind; thus, conferring resistance. Similarly, 85–90% of isoniazid (INH)-resistant isolates can be detected by sequencing the fabG1-inhA promoter region, fabG1, and katG (4,5). INH resistance is primarily attributed to mutations in the fabG1-inhA promoter region which leads to overproduction of the drug target and mutations within katG which inhibits activation of INH. Rapid detection of the presence of these mutations in rpoB, fabG1-inhA promoter, fabG1, and katG can indicate that the isolate is resistant to RIF or INH weeks before growth-based DST results would typically be available.
Though the genetic basis of resistance for many anti-tuberculosis drugs is well understood (4,6), there are still gaps in our knowledge. For example, a clear interpretation for a specific mutation may not be defined, especially for those that are rare or novel. Additionally, sometimes discordance between growth-based and molecular methods may be observed. One example is isolates with specific clinically relevant rpoB mutations that are associated with low-level rifampin resistance as isolates with these mutations may test as susceptible to rifampin in growth-based DST. Similarly, some isolates that test resistant by growth-based methods could have mutations in genetic loci other than those examined. Additionally, for new and repurposed drugs (e.g., clofazimine, bedaquiline, and linezolid), we are continuing to learn more about mechanisms of resistance. Thus, the interpretation of both DNA sequencing results and results from growth-based methods must be considered in context with specific criteria and test limitations for each individual drug based on existing data. Although discordance can occur, accuracy of detection of drug resistance is improved when considering MDDR and growth-based DST results in combination.
- Specific limitations and considerations for interpretation differ by drug. A helpful resource for understanding how resistance develops in MTBC and how molecular testing for MTBC is performed can be obtained through the Association of Public Health Laboratories (https://www.aphl.org/programs/infectious_disease/tuberculosis/Pages/Training-Modules.aspx). WHO has an available resource to aid interpretation of mutations conferring resistance (7).
- Additional resources for medical consultation are available from your state TB program or the TB Centers of Excellence for Training, Education, and Medical Consultation (877-390-6682)
DNA sequencing of genetic loci known to contribute to antibiotic resistance in MTBC will be performed using primarily a tNGS assay (8). This assay examines specific genomic regions. Briefly, DNA regions of interest are amplified by polymerase chain reaction (PCR), and sequencing libraries are generated using the Illumina Nextera XT Library Prep Kit. The libraries are sequenced on the Illumina MiniSeq platform for the MDDR Service. Thousands of reads are generated for each genetic locus that is sequenced and the reads are mapped to a reference sequence and analyzed for genetic differences (i.e., mutations).
One benefit of tNGS is a substantial increase in data generated compared to conventional DNA sequencing methods like pyrosequencing and Sanger sequencing technologies previously utilized for the CDC MDDR service. Pyrosequencing and Sanger sequencing will be retained for special circumstances including for testing of DNAs extracted from fixed tissue samples from CDC’s Infectious Diseases Pathology Branch.
In addition to sequencing, all isolates, including those grown from sediments, will undergo growth-based DST. Material from nucleic acid amplification test (NAAT) positive sediments will be inoculated into growth medium to obtain an MTBC isolate. Growth-based DST will be performed using the agar proportion method. Pyrazinamide testing will be performed by the MGIT 960 method.
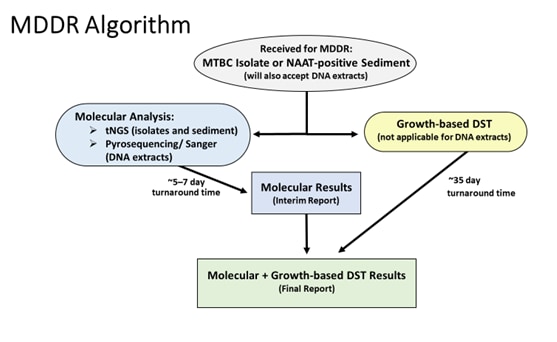
Figure 2. The testing algorithm for the Molecular Detection of Drug Resistance (MDDR) service is shown for samples received. The anticipated mean turnaround time for both molecular analysis and growth-based drug susceptibility testing (DST) is indicated. An interim report including only the molecular results will be provided followed by a final report including growth-based DST results, when applicable. DNA extracts are only accepted from CDC’s Infectious Diseases Pathology Branch.
tNGS: targeted next generation sequencing assay, NAAT: nucleic acid amplification testing
The tNGS panel was designed to detect resistance-associated mutations for first- and second-line antituberculosis drugs, as well as new and repurposed drugs like bedaquiline, linezolid, and clofazimine.
Table 1. Description of information regarding genetic loci examined in targeted next generation sequencing assay (tNGS), the associated antituberculosis drug, as well as the position and types of mutations that will be included in the laboratory report
| Drug | Genetic locus | Upstream region1 | Nucleotide position in rRNA2 | Codons3 | Rubric for Laboratory Reporting4 |
|---|---|---|---|---|---|
| Rifampin | rpoB rifampin- resistance- determining region (RRDR) |
Gly426 to Leu452 | All mutations in the RRDR and codons 170 and 491 reported | ||
| rpoB codon 170 | Val170 | ||||
| rpoB codon 491 | Ile491 | ||||
| Isoniazid | katG | Val1 to 741* | All mutations except lineage markers and synonymous mutations at positions other than codon 1 | ||
| fabG1- inhA_upstream | -140 to -1 | All mutations upstream of fabG1 start codon and mutations at codon 203 only | |||
| fabG1 codon 203 | Leu203 | ||||
| Ethambutol | embB (partial) | Thr277 to Thr437 | All mutations except lineage markers and synonymous mutations | ||
| Pyrazinamide | pncA | -40 to -1 | Met1 to 187* | All mutations upstream of the pncA start codon and all mutations in the open reading frame5 except synonymous mutations | |
| Fluoroquinolones | gyrA quinolone- resistance– determiningregion (QRDR) | Gly88 to Asp94 | All mutations at codons 88 to 94 except synonymous mutations | ||
| gyrB | Arg446 to Gly537 | All mutations except synonymous mutations | |||
| Amikacin Capreomycin Kanamycin | rrs (partial) | 1177 to 1537 | Mutations at nucleotides 1401, 1402, and 1484 only | ||
| Kanamycin | eis_upstream | -127 to -1 | All mutations | ||
| Bedaquiline | atpE | -48 to -1 | Met1 to 82* | All mutations upstream of the atpE start codon and non-synonymous mutations in the open reading frame5 | |
| rv0678 | -84 to -1 | Val1 to 166* | All mutations upstream of the rv0678 start codon and non- synonymous mutations in the open reading frame5 | ||
| pepQ | -33 to -1 | Val1 to 373* | All mutations upstream of the pepQ start codon and non-synonymous mutations in the open reading frame5 | ||
| Clofazimine | rv0678 | -84 to -1 | Val1 to 166* | All mutations upstream of the rv0678 start codon and non- synonymous mutations in the open reading frame5 | |
| pepQ | -33 to -1 | Val1 to 373* | All mutations upstream of the pepQ start codon and non-synonymous mutations in the open reading frame5 | ||
| Linezolid | rplC | -18 to -1 | Met1 to 217* | All mutations upstream of the rplC start codon and non-synonymous mutations in the open reading frame5 | |
| rrl (partial) | 2003 to 2367 and 2449 to 3056 | All mutations |
1 Coordinates are relative to the predicted start codon.
2 Nucleotide positions are indicated for rrs and rrl because a promoter region and codon numbering are not applicable for these rRNA genes.
3 Codon numbers define amino acid position in the translated protein.
4 When present in the open reading frame, mutations will be reported as indicated with the exclusion criteria defined. When absent, the report will indicate no mutation detected. Mutations are identified by comparing sequencing data for each sample to the sequence of the H37Rv reference isolate. The minimum reportable alternate allele frequency threshold for the analytic pipeline results is 10%.
5 Open reading frame is the genetic sequence that is transcribed into mRNA and ultimately translated into protein corresponding with the codon position defined for some of the genetic loci examined in the tNGS assay.
* Indicates a STOP codon.
A validation was completed with comparison of results from the tNGS assay with those from Sanger sequencing. These data showed that for reportable mutations in which the same genetic regions and type of mutations were assessed for each method, the results matched >99% of the time indicating that the tNGS assay is as good as Sanger sequencing. Additionally, the tNGS assay includes the examination of expanded genetic loci relative to Sanger sequencing and improves the ability to detect minor variant populations (≥10% of population) for new drugs and improves the performance of MDDR for previously evaluated drugs such as INH. The performance characteristics of MDDR based on Sanger sequencing results obtained through the service from 2012 to 2021 are included below using growth-based DST (agar proportion for all drugs with exception of MGIT for pyrazinamide) result (either susceptible or resistant) as the reference standard. This table will be updated with tNGS results at least biennially and include data for new drugs (e.g., bedaquiline, linezolid, and clofazimine), as applicable.
Table 2. Performance characteristics of MDDR by Drug Using Results from Sanger
Sequencing 2012−2021
| Drug | Locus or loci examined
|
Sensitivity (%) | Specificity (%) |
| Rifampin | rpoB RRDR1, codons 170 and 491
|
99.8 | 91.82 |
| Isoniazid | fabG1-inhA_upstream, katG codon 315, fabG1 codon 203 | 93.6 | 99.2 |
| Ethambutol | embB (partial) | 80.6 | 94.2 |
| Pyrazinamide | pncA | 69.83 | 95.7 |
| Fluoroquinolones | gyrA QRDR4 | 86.4 | 99.3 |
| Kanamycin
|
rrs (partial)
eis_upstream |
93.9 | 99.3 |
| Amakacin | rrs (partial)
|
95.8 | 99.9 |
| Capreomycin | rrs (partial)
tlyA |
98.3 | 95.3 |
Rifampin resistance determining region
2 Specificity for rifampin resistance likely impacted by challenges with detection of growth-based resistance in isolates with mutations resulting in low-level rifampin resistance.
3 Some isolates without detected pncA mutations tested as resistant by MGIT (i.e., growth-based method).
4 Quinolone resistance determining region
The limitations of the MDDR service can be attributed primarily to gaps in knowledge and the LOD of the assays being used. Another limitation is that the clinical relevance of some mutations remains unknown. When mutations detected are known to confer resistance, MTBC can be confidently called resistant. However, when mutations are novel, or there is insufficient data to definitively establish an interpretation, resistance can be neither confirmed nor ruled out. Likewise, if no mutation is detected, resistance cannot always be ruled out due to the possible presence of mutations in regions of the chromosome that are not covered by the sequencing assay.
Genetic analysis may miss heteroresistance (i.e., only a portion of organisms in the population being tested carry a mutation associated with resistance that is below LOD of the assay). Used alone, MDDR and growth-based DST are imperfect, yet when used in combination, accuracy of the detection of drug resistance is improved.
MDDR is listed in the CDC Test Directory as CDC-10186 (https://www.cdc.gov/laboratory/specimen-submission/detail.html?CDCTestCode=CDC-10186) and pre-approval (see Section C. 3.) is required for this test. Isolates of MTBC and MTBC NAAT+ sediments may be submitted by U.S. Public Health Laboratories for MDDR for the following reasons:
- By patient history, there is a high-risk of RIF resistance or multidrug-resistant (MDR) TB (including those previously treated for TB, a contact of a drug resistant TB case, from an area with high rates of drug resistance, or exhibiting a lack of clinical response to therapy)
- Known RIF resistance (by molecular test or growth-based DST)
- Patients where the result of drug resistance will predictably have a high public health impact (e.g., daycare workers, nurses)
- Patient is unable to tolerate first-line drugs (e.g., allergy to RIF)
- Mixed or non-viable cultures (i.e., growth-based DST not possible)
- Isolates which fail to grow in DST medium
- Other situations considered on a case-by-case basis
-
- Please contact TBLab@cdc.gov to discuss other circumstances where testing may be needed.
Note: Submitters must retain the original isolate/culture/inoculum in the submitting laboratory for the duration of testing at CDC.
Note: MDDR does not take the place of conventional culture and DST for MTBC. Standard of care testing (i.e., smear, culture, and DST) should be performed in the submitting laboratory when MTBC NAAT+ sediments are submitted for MDDR testing. Results of MDDR should not be used to rule out the presence of MTBC in a sample when no amplification is detected by MDDR.
Acceptable Sample Types
- MTBC NAAT+ sediment
- Pure MTBC isolate on solid medium or in broth medium
- Mixed cultures known to contain MTBC
Only one sample per patient should be submitted; however, testing of duplicate samples will be considered on a case-by-case basis. Contact the CDC MDDR Point of Contact at TBLab@cdc.gov for approval.
Acceptable Collection, Storage and Preservation Prior to Shipping
- MTBC NAAT+ sediments: Stored at 2 to 8°C up to 30 days post collection or -20°C or lower up to 60 days post collection
- MTBC isolates: Stored at 2 to 8°C (liquid media up to 120 days post collection; solid media up to 180 days post collection); 15 to 25°C (liquid media up to 120 days post collection; solid media up to 180 days post collection); -20°C or lower up to 120 days post collection (liquid media); -70°C or lower up to 5 years post collection (liquid media)
Minimum Volume Required for Liquid Samples
- MTBC NAAT+ sediment: Submit at least 0.5 ml, 1 ml preferred
- MTBC isolates in liquid medium: Submit at least 0.5 ml, 1 ml preferred
Acceptable Transport Medium
- Solid media (i.e., Middlebrook 7H10 or 7H11 plates/slants, Lowenstein-Jensen [LJ] slants)
- Liquid media (i.e., Middlebrook 7H9, Mycobacterial Growth Indicator Tube [MGIT], BacT/ALERT, VersaTREK) or MTBC growth from solid media, suspended in saline or sterile water
Required Specimen Labeling
Test is subject to CLIA regulations and requires two primary patient identifiers (e.g., patient first and last name, date of birth or both, sex and age, unique patient identifier from time of collection, such as medical record number) on the specimen container and on the test requisition.
Submitters should complete all portions of the MDDR request form (http://www.cdc.gov/tb/topic/laboratory/mddrsubmissionform.pdf) and submit via email (TBLab@cdc.gov) or through the CDC Specimen Test Order and Reporting (CSTOR) web portal. CSTOR allows submitters to request and obtain approval to send samples for testing, complete the CDC requisition form (50.34), validate specimen data, and securely view and download test reports (https://www.cdc.gov/laboratory/specimen-submission/specimen-test-order-reporting.html). Submitters are encouraged to use CSTOR for MDDR requests. Once approved, CDC will provide submission instructions. Please attach the MDDR request form to the CDC requisition (https://www.cdc.gov/laboratory/specimen-submission/form.html) or manifests, if using CSTOR. Please note that specimen collection date, date sent to CDC, and material submitted (i.e., original material if sediment is submitted or isolate) are required fields on the CDC requisition (50.34) for submission.
Shipping address:
TB Lab/ Atanaska Marinova-Petkova
Centers for Disease Control and Prevention
RDSB/STATT Unit 29
1600 Clifton Road, NE Atlanta, GA 30329
404-718-5254 or 404-639-2455
Questions regarding the use of the Association of Public Health Laboratory (APHL)
FedEx Account for shipment should be addressed to APHL (sarah.buss@aphl.org; erin.estes@aphl.org ).
Shipping Instructions which include Specimen Handling Requirements
- For liquid sample, submit in a screwcap cryovial that has been sealed with parafilm. Do not send any samples in 15- or 50-mL conical tubes.
- Sediments and isolates should be shipped at room temperature and must be received within 5 days of shipment.
- Sediments and isolates should be shipped via overnight service to CDC Monday through Thursday. Do not ship on Friday. Samples should be shipped in compliance with federal regulation. MTBC isolates must be shipped as Infectious substances (Div. 6.2, Class Category A).
CDC shipping information link: https://www.cdc.gov/laboratory/specimen- submission/index.html
A Specimen Packing and Shipping Guidance Fact Sheet can be found on the CDC internet site under Submitting Specimens to CDC/Shipping and Packing page (CDC Fact Sheet).
Reports will be sent via CDC’s Laboratory Information Management System (LIMS) to the submitting public health laboratory when the DNA sequencing results are available. A final report will be issued through the LIMS when growth-based DST results are available. Comments regarding discordance between growth-based DST results and DNA sequencing, if appropriate, will be included. The submitting public health laboratory is responsible for further dissemination of CDC reports to the TB program and healthcare provider, as appropriate.
Laboratorians, TB control personnel, and healthcare providers can contact the CDC for help in interpretation of reports.
e-mail: TBLab@cdc.gov
Telephone: 404-639-2455
D. References
- Campbell P. J., G. P. Morlock, R.D. Sikes, T.L. Dalton, B. Metchock, A.M. Starks, D. P. Hooks, L. S. Cowan, B.B. Plikaytis, and J.E. Posey. 2011. Molecular Detection of Mutations Associated with First and Second-Line Drug Resistance Compared with Conventional Drug Susceptibility Testing in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy. 55(5):2032-41.
- Association of Public Health Laboratories. Issues in Mycobacterium tuberculosis complex (MTBC) Drug Susceptibility Testing: Rifampin (RIF). April 2019. (Issues in Mycobacterium tuberculosis Complex (MTBC) Drug Susceptibility Testing: Rifampin (RIF) (aphl.org))
- Andre E., L. Goeminne, A. Cabibbe, P. Beckert, B. Kabamba Mukadi, V. Mathys, S. Gagneux, S. Niemann, J. Van Ingen, E. Cambau. 2017. Consensus Numbering System for the Rifampicin Resistance-associated rpoB gene mutations in pathogenic mycobacteria. Clinical Microbiology and Infection. 23(3): 167-172.
- Miotto P., Y. Zhang, D.M. Cirrillo, W.C. Yam. 2018. Drug Resistance Mechanisms and Drug Susceptibility Testing for Tuberculosis. Respirology. 23(12): 1098-1113.
- Kandler J.L., A.D. Mercante, T.L. Dalton, M. N. Ezewudo, L.S. Cowan, S.P. Burns, B. Metchock, Global PETTS Investigators, P. Cegielski, and J.E. Posey. Validation of Novel Mycobacterium tuberculosis Isoniazid Resistance Mutations Not Detectable by Common Molecular Tests. 2018. Antimicrobial Agents and Chemotherapy. 62(10):e00974-18. doi: 10.1128/AAC.00974-18.
- Allix-Béguec C, Arandjelovic I, Bi L, Beckert P, Bonnet M, Bradley P, Cabibbe AM, Cancino-Muñoz I, Caulfield MJ, Chaiprasert A, Cirillo DM, et al. Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing. New England Journal of Medicine. 2018. 11;379(15):1403-1415.
- World Health Organization. Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. 2021. https://www.who.int/publications/i/item/9789240028173
- World Health Organization and Foundation for Innovative New Diagnostics. Technical Guide on Next-generation Sequencing Technologies for the Detection of Mutations Associated with Drug Resistance in Mycobacterium tuberculosis complex. 2018. WHO-CDS-TB-2018.19-eng.pdf