Enteric Diseases Laboratory Branch
CDC’s lead laboratory supporting the effort to prevent illness caused by enteric (intestinal) diseases.

Activities
- Detect, investigate, and track trends in foodborne disease outbreaks alongside epidemiologists. Conduct research about foodborne pathogens and antimicrobial susceptibility and improve molecular and phenotypic detection methods
- Build capacity for molecular surveillance of foodborne infections in the United States and internationally
- Provide innovative laboratory tools and quality systems to identify, subtype, and enhance surveillance to minimize impacts of foodborne illness
- Generate, mobilize, and leverage scientifically reliable data to help reduce the burden of antimicrobial-resistant enteric bacteria on human, animal, and environmental health
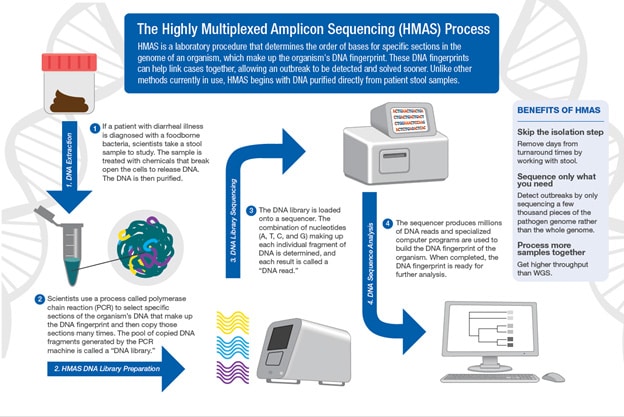
Bioinformatics and Metagenomics Team
The Bioinformatics and Metagenomics Team (BIOME) provides bioinformatics support and research for foodborne pathogen surveillance and outbreak investigation. The team also provides bioinformatics analyses for bacterial outbreak detection, public health laboratory support, and development of new bioinformatics tools.
BIOME accomplishes these goals by:
- Supporting PulseNet to connect data from DNA fingerprinting of bacteria that make people sick
- Using direct-from-stool techniques like highly multiplexted amplicon sequencing (HMAS)
- Developing metagenomics shotgun sequencing for foodborne illness outbreak surveillance
These techniques will allow public health laboratories to continue and improve foodborne illness surveillance as clinical laboratories adopt culture independent diagnostic testing.
National Antimicrobial Resistance Surveillance Team
The National Antimicrobial Resistance Surveillance Team’s (NARST) vision is to contribute to a wholistic understanding of the public health burden of antimicrobial-resistant bacteria in food and the environment causing intestinal diseases.
The National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) was established in 1996 and is a collaboration among state and local public health departments, CDC, the U.S. Food and Drug Administration (FDA), and the U.S. Department of Agriculture (USDA). NARMS surveillance in all 50 states helps identify patterns of emerging resistance that can guide public health awareness and policy efforts to protect people from resistant infections. This information is of interest to many groups, including federal regulatory agencies, policymakers, consumer advocacy groups, industry, and the public.
NARST, the laboratory component of NARMS-CDC, tracks antimicrobial resistance and studies patterns of emerging resistance in intestinal bacteria transmitted commonly through food, people, animals, and the environment. The laboratory also performs research to understand the genes responsible for resistance, the ways in which resistance is spread, and how antimicrobial-resistant infections differ from antimicrobial-susceptible infections.
NARST accomplishes these goals for surveillance on sporadic cases, outbreaks of illness, and special studies by conducting susceptibility testing and sequencing whole pathogen genomes.
National Botulism Laboratory
The National Botulism Laboratory’s (NBL) vision is a safer world through botulism investigations and applied research.
NBL maintains CDC expertise for laboratory investigations of botulism. It provides technical assistance to U.S. and international public health laboratories by detecting and serotyping botulinum neurotoxins. It also isolates and identifies botulinum-toxin-producing species of Clostridium.
NBL develops and validates new detection and identification methods to prepare for potential bioterrorism events involving botulinum neurotoxin. These methods can also be used in response to unintentional events. NBL also uses next-generation sequencing methods to characterize and subtype Clostridium botulinum and related organisms, including a national molecular surveillance database for comparing genetic relatedness among strains across the United States and identifying trends among botulism cases.
NBL’s mission is to provide subject matter expertise and maintain emergency preparedness by:
- Providing exceptional reference laboratory testing services
- Developing and validating new laboratory tests
- Investigating the diversity of botulinum neurotoxin and botulinum neurotoxin–producing bacterial strains of Clostridium
PulseNet Reference Outbreak Surveillance Team
PulseNet is a national laboratory network that connects foodborne illness cases to detect outbreaks. PulseNet uses DNA fingerprinting of bacteria making people sick to detect thousands of local and multistate outbreaks. PulseNet International performs a similar role for foodborne illnesses globally.
The PulseNet Reference Outbreak Surveillance Team is composed of several laboratories and units that conduct a broad range of activities to support pathogen discovery and characterization, foodborne outbreak surveillance and detection, whole genome sequencing, laboratory capacity building, and global public health partnerships.
To sustain our ability to provide quality service, these reference laboratories conduct applied research to better understand the biology, pathogenesis, and epidemiology of enteric bacterial pathogens and to develop improved diagnostic methods to detect and characterize them:
- National Campylobacter and Helicobacter Reference Laboratory
- National Salmonella Reference Laboratory
- National Listeria, Yersinia, Vibrio, and Enterobacterales Reference Laboratory
- National Escherichia and Shigella Reference Laboratory