How does MicrobeNet work?
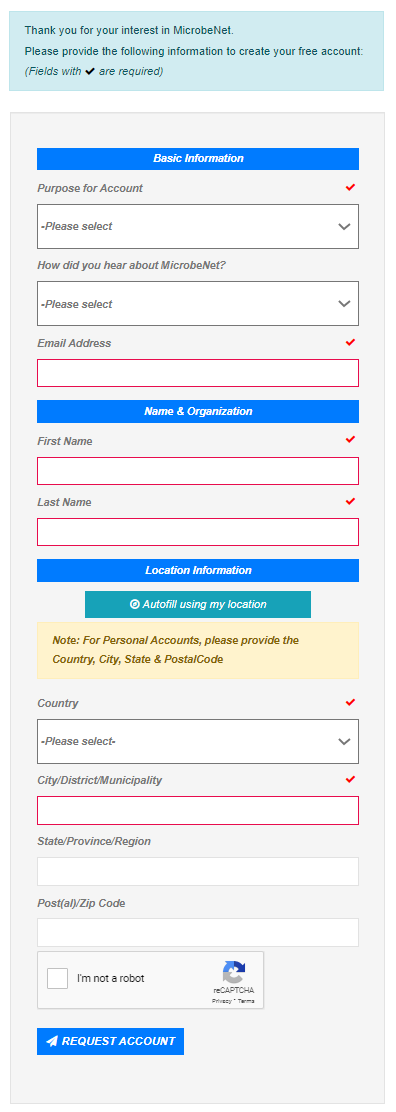
New users can request a free account by completing the registration form. All you need is internet access and an email address.
Registered users can access MicrobeNet by following a few simple steps:
- Step 1: Sign in at microbenet.cdc.gov
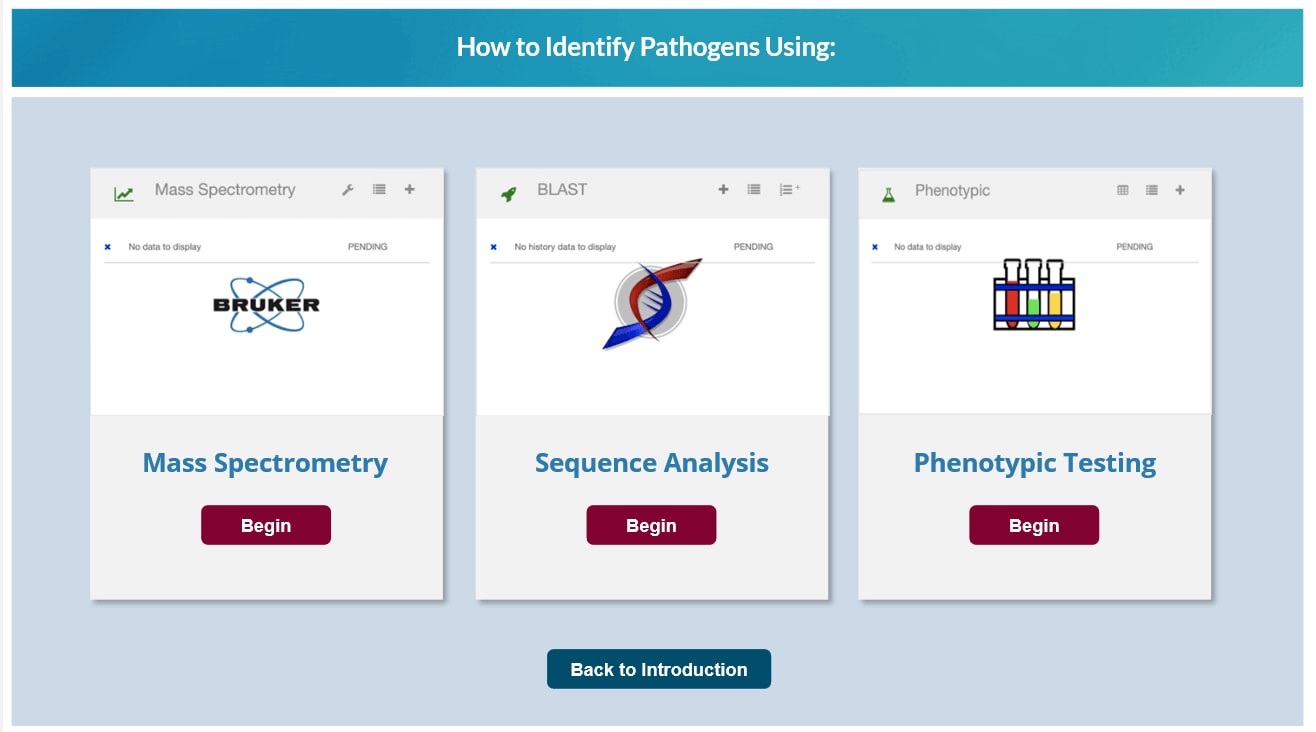
- Step 2: Submit your 16S and ITS gene sequences, MALDI-TOF spectra, and/or biochemical test panel results.
- Step 3: MicrobeNet returns the closest identified matches within minutes.
- Step 4: Click on the results to find out more information about the identified pathogen.
- Step 5: Still have questions? Contact a CDC subject matter expert specific to the identified pathogen by using the contact information listed in MicrobeNet.
Save time and resources by not creating redundant databases and avoiding unnecessary shipment of samples to CDC laboratories for identification.
Learn how to use MicrobeNet with our training videosexternal icon.
Page last reviewed: February 24, 2022