Prospecting for Zoonotic Pathogens by Using Targeted DNA Enrichment
Egie E. Enabulele, Winka Le Clec’h, Emma K. Roberts, Cody W. Thompson, Molly M. McDonough, Adam W. Ferguson, Robert D. Bradley, Timothy J. C. Anderson, and Roy N. Platt
Author affiliations: Texas Biomedical Research Institute, San Antonio, Texas, USA (E.E. Enabulele, W. Le Clec’h, T.J.C. Anderson, R.N. Platt II); Texas Tech University, Lubbock, Texas, USA (E.K. Roberts, R.D. Bradley); University of Michigan, Ann Arbor, Michigan, USA (C.W. Thompson); Chicago State University, Chicago, Illinois, USA (M.M. McDonough); Field Museum of Natural History, Chicago (A.W. Ferguson)
Main Article
Figure 3
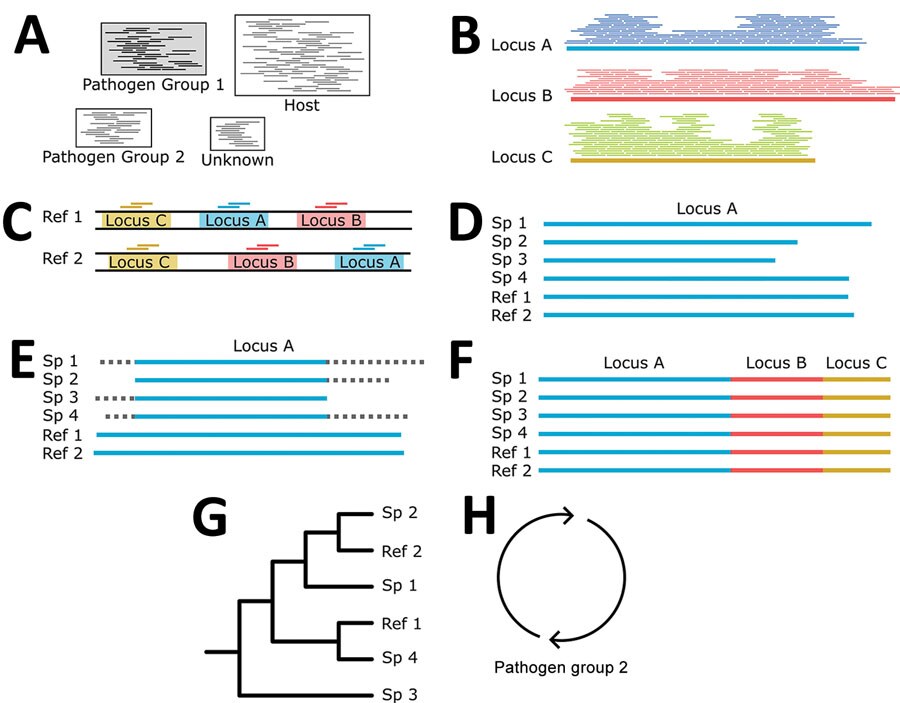
Figure 3. Building phylogenies from parasite reads for study of prospecting for zoonotic pathogens by using targeted DNA enrichment. A) After read classification, we extracted all the reads associated with a pathogen group. B) Those reads were assembled into contigs with a genome assembler. C) Simultaneously, we identified and extracted the target loci from all members of the pathogen group with available reference genomes to ensure that our final phylogeny has representatives from as many members of the pathogen group as possible. D, E) For each targeted locus, we combined the assembled contigs (D) and genome extracted loci for (E) multiple sequence alignment and trimming. F, G) Each aligned and trimmed locus is concatenated together (F) for phylogenetic analyses (G). H) If necessary, those steps are repeated for reads classified in other pathogen groups. Ref, reference; Sp, specimen.
Main Article
Page created: June 07, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
