Volume 30, Number 4—April 2024
Research
Crimean-Congo Hemorrhagic Fever Virus Diversity and Reassortment, Pakistan, 2017–2020
Figure 4
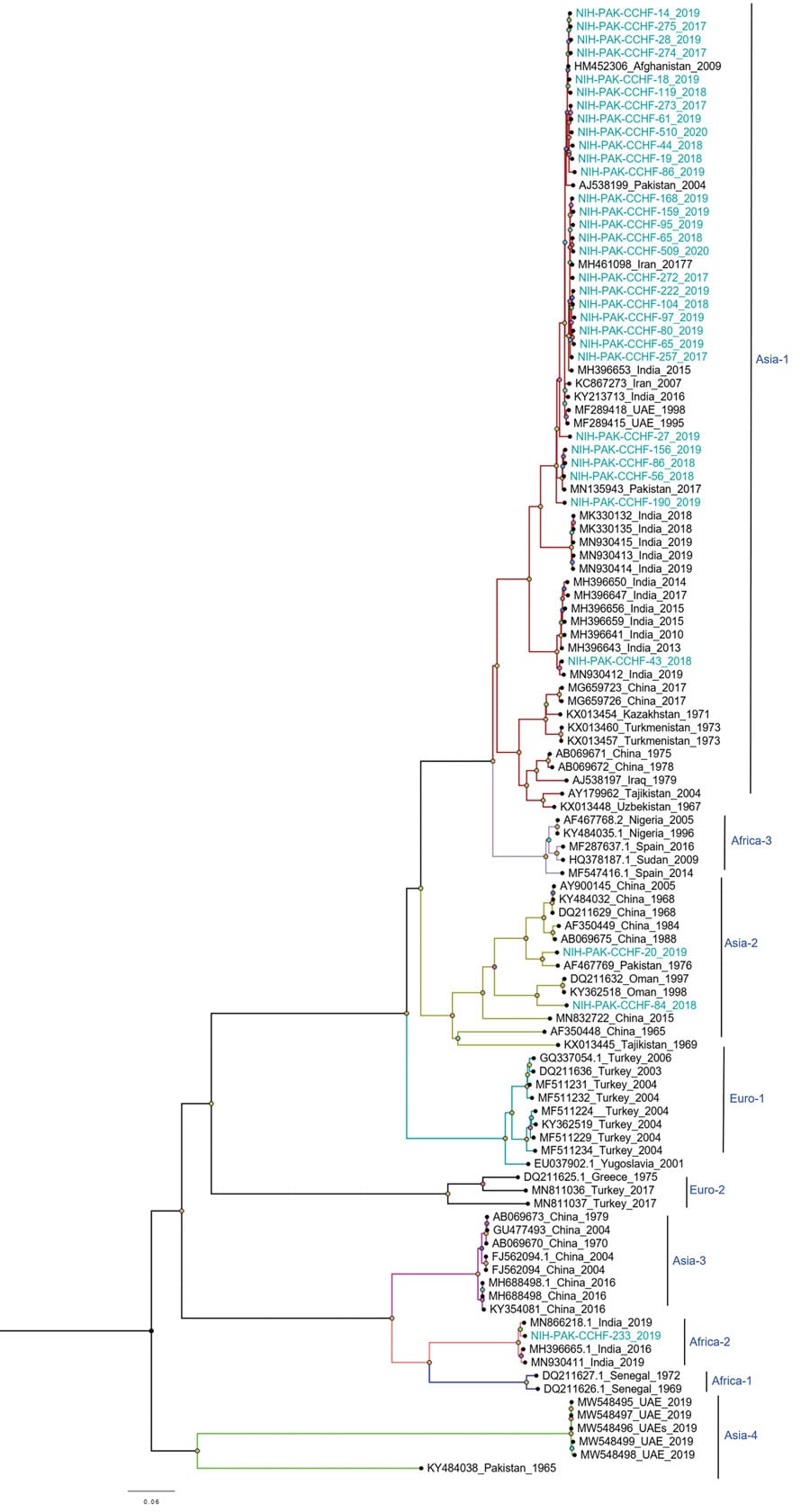
Figure 4. Phylogenetic analysis of full-length medium gene segments of Crimean-Congo hemorrhagic fever virus in study of virus diversity and reassortment, Pakistan, 2017–2020. Midpoint-rooted trees were generated by using the maximum-likelihood method. Blue-green text indicates sequences from this study. Most M segments from Pakistan clustered with the Asia-1 genotype/clade, but 3 reassorted sequences clustered with the Asia-2 clade, and 1 reassorted sequence clustered with the Africa-2 clade. Scale bar indicates nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: January 31, 2024
Page updated: March 20, 2024
Page reviewed: March 20, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.