COVID-19 Science Update released: June 2, 2020 Edition 18

The COVID-19 Science Update summarizes new and emerging scientific data for public health professionals to meet the challenges of this fast-moving pandemic. Weekly, staff from the CDC COVID-19 Response and the CDC Library systematically review literature in the WHO COVID-19 databaseexternal icon, and select publications and preprints for public health priority topics in the CDC Science Agenda for COVID-19 and CDC COVID-19 Response Health Equity Strategy.
Here you can find all previous COVID-19 Science Updates.
PEER-REVIEWED
Prevalence and clinical presentation of health care workers with symptoms of coronavirus disease 2019 in 2 Dutch hospitals during an early phase of the pandemicexternal icon. Kluytmans-van den Bergh et al. JAMA Network (May 21, 2020).
Key findings:
- 6% of symptomatic healthcare workers (HCW) at two Dutch hospitals tested positive for SARS-CoV-2 in early March (86/1,353).
- One-quarter (21/86) of those infected had no direct patient contact.
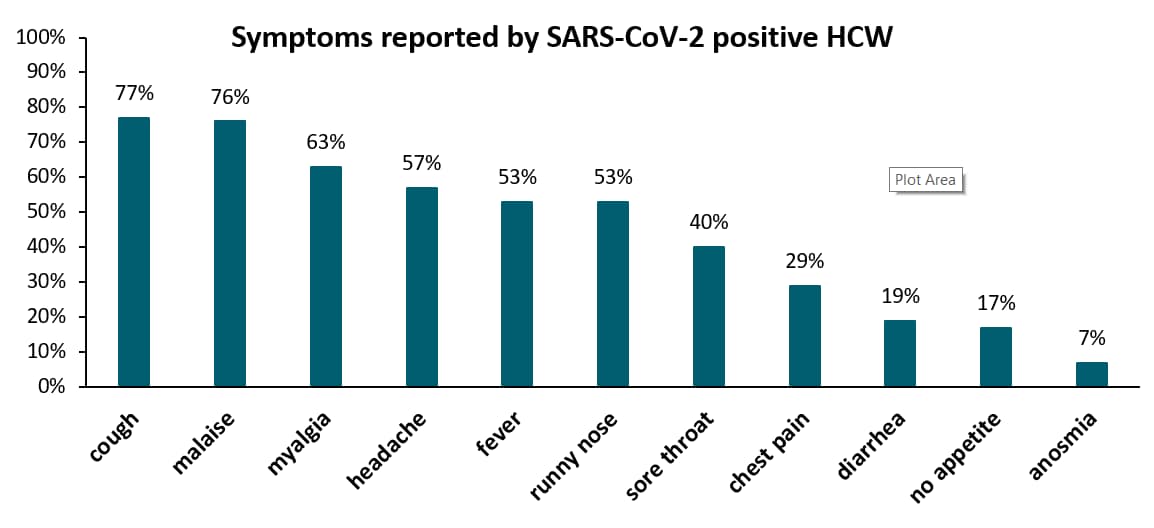
- Symptoms were mild; most frequently reported symptoms were cough and fatigue (Figure).
Methods: 9,705 HCW at two hospitals were screened for symptoms between March 7 and 12, 2020; SARS-CoV-2 RT-PCR testing of OP swabs was performed for workers reporting fever or respiratory symptoms in the last 10 days. Limitations: Likely an underestimate since asymptomatic workers were not tested.
Implications: HCW who test positive for SARS-CoV-2 can have mild symptoms. SARS-CoV-2 screening of HCW can support prevention and control measures in hospitals.
Figure:
Note: Adapted from Kluytmans-van den Bergh et al. Proportion of SARS-CoV-2 positive HCW self-reporting symptoms of COVID-19. Licensed under CC-BY.
Self-quarantine and weight gain related risk factors during the COVID-19 pandemicexternal icon. Zachary et al. Obesity Research & Clinical Practice (May 15, 2020).
Key findings:
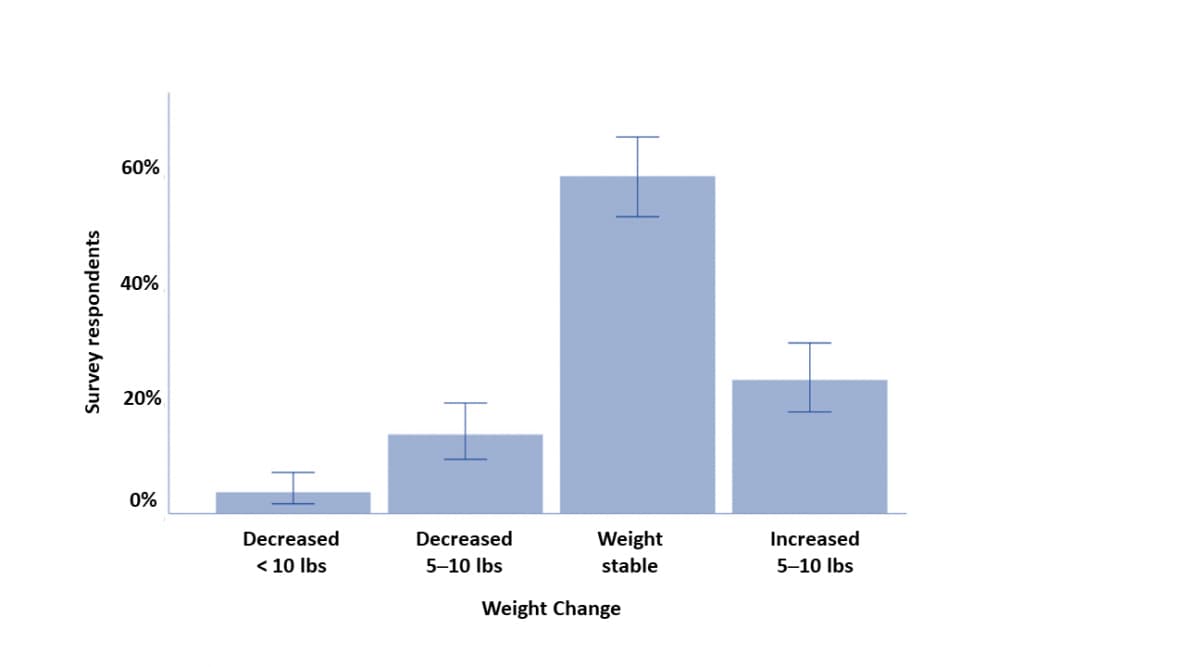
- During self-quarantine, 22% of people reported gaining 5 to 10 pounds (Figure).
- People who gained weight reported eating more in response to stress.
Methods: Online survey completed by people in self-quarantine with questions on demographics, social networks, weight & lifestyle inventory, sedentary behavior, physical activity, and perceived stress. Research announcement distributed to 1,200 adults on Facebook; 173 (14%) participated. Limitations: Sample may not be representative and results not generalizable; no details on inclusion criteria for who received the invite; self-reported outcomes.
Implications: Weight gain from stress may occur during COVID-19 self-quarantine.
Figure:
Note: Adapted from Zachary et al. Reported weight change during COVID-19 self-quarantine. Error bars represent 95% CI. This article was published in Obesity Research & Clinical Practice, Vol 14, Zachary et al., Self-quarantine and weight gain related risk factors during the COVID-19 pandemic, Page 210-216, Copyright Asia Oceania Association for the Study of Obesity 2020. This article is currently available at the Elsevier COVID-19 resource center: https://www.elsevier.com/connect/coronavirus-information-centerexternal icon.
Although many risk factors for COVID-19 infection and disease severity have been well-established in the literature, few studies have addressed disparities in COVID-19 disease outcomes by race and ethnicity. Identifying factors contributing to disparities is critical for developing public health intervention measures.
PEER-REVIEWED
A. Disparities in outcomes among COVID-19 patients in a large health care system in Californiaexternal icon. Azar et al. Health Affairs (May 21, 2020).
Key findings:
- African American patients with COVID-19 were over 2.5 times more likely to be hospitalized than non-Hispanic white patients after adjusting for age, sex, and select clinical comorbidities.
- In models that included sociodemographic factors (insurance type, income), the increased odds were slightly attenuated
Methods: 1,052 adults with confirmed COVID-19 disease who had at least one encounter at a San Francisco Sutter Healthcare clinic or hospital during first three months of 2020. Patient-level data were extracted from electronic health records. Limitations: One geographical area; income was assigned at ZIP-code level; the race category for African American appears to have included Hispanic blacks; data in appendices not available for review.
B. Hospitalization and mortality among black patients and white patients with COVID-19external icon. Price-Haywood et al. NEJM (May 27, 2020).
Key findings:
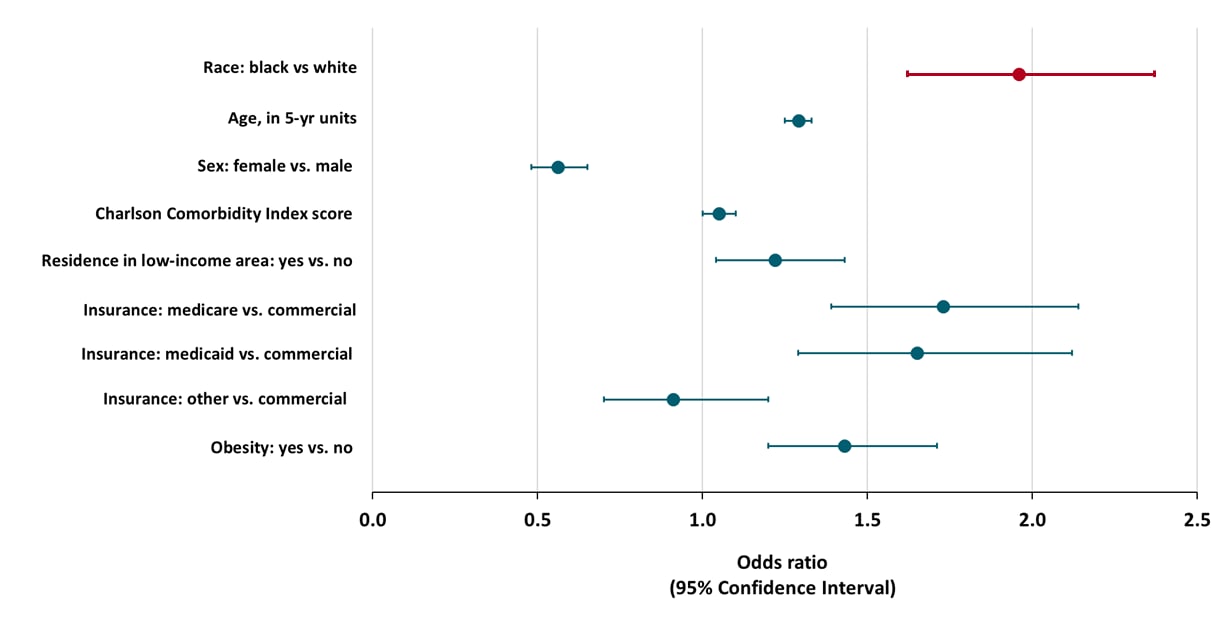
- Non-Hispanic Black patients with COVID-19 were twice as likely to be hospitalized than non-Hispanic White patients after adjusting for age, sex, comorbidities, residence in a low-income area, insurance type, and obesity (Figure).
- In-hospital mortality was not significantly different between non-Hispanic Black and non-Hispanic White patients.
Methods: Retrospective cohort study of 3,481 confirmed COVID-19 patients attending the Ochsner Health system in Louisiana, USA, between March 1 and April 11, 2020. In-hospital deaths assessed through May 7, 2020. Limitations: One geographical area.
Figure:
Note: Adapted from Price-Haywood et al. Odds Ratios for hospitalization among COVID-19 patients. Model includes race, as well as age, sex, Charlson Comorbidity Index score, residence in a low-income area, insurance plan, and obesity. From NEJM 2020; 382:2534-2543. DOI: 10.1056/NEJMsa2011686. Copyright © 2020 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
Implications for 2 studies (Azar et al. and Price-Haywood et al.): Non-Hispanic Black patients are more vulnerable to SARS-CoV-2 infection and compromise a disproportionate fraction of deaths from COVID-19 than non-Hispanic White patients. COVID-19 illness may be more severe for non-Hispanic Black persons than non-Hispanic white persons; analyses must carefully control for other factors related to disease severity. Analyses are also needed that examine infection risk and illness severity among Hispanic, Asian, American Indian/Alaska Native and other racial or ethnic minority persons.
PEER-REVIEWED
Metabolic associated fatty liver disease increases COVID-19 disease severity in non-diabetic patients.external icon Gao et al. Gastroenterology & Hepatology (May 21, 2020).
Key findings:
- In non-diabetic patients with COVID-19, having metabolic-associated fatty liver disease (MAFLD) was associated with 4 times increased risk of severe COVID-19.
- The association between MAFLD and severe COVID-19 was independent of age, sex, smoking status, obesity, hypertension, and high cholesterol.
Methods: Case-control study among 130 hospitalized COVID-19 patients in China. Severe disease defined as respiratory distress or failure. MAFLD identified by CT scan and subsequently diagnosed using a clinical case definition. Limitations: May not be generalizable to U.S. population.
Implications: MAFLD is a risk factor for severe COVID-19. Early identification of MALFD in COVID-19 patients may inform clinical decision making.
Universal testing of patients and their support persons for COVID-19 when presenting for admission to labor and delivery within the Mount Sinai Health Systemexternal icon. Buckley et al. American Journal of Obstetrics and Gynecology MFM (May 22, 2020).
Key findings:
- 16% of asymptomatic women in labor (50/307) and 20% of their support companions (39/199) tested positive for SARS-CoV-2.
- Among the 171 COVID-19 negative women, 9% had companions that tested positive for SARS-CoV-2.
Methods: 506 women in labor and their companions were universally screened for SARS-CoV-2 at two institutions within Mount Sinai Health System, New York City, NY between April 4 and 15, 2020. Limitations: May not be generalizable to other hospitals, cities, and time periods.
Implications: SARS-CoV-2 screening of patients and their companions can support prevention control measures in hospitals.
Remdesivir is an antiviral drug that has previously been shown as effective against human coronaviruses in laboratory and animal models. However, evidence related to the effectiveness of remdesivir for treating COVID-19 in humans is uncertain. Currently, there are nearly 30 clinical trials planned or underway to test remdesivir as a treatment for COVID-19. The following articles report results from two such trials.
PEER-REVIEWED
A. Remdesivir for the treatment of COVID-19 — preliminary reportexternal icon. Beigel et al. NEJM (May 22, 2020).
Key findings:
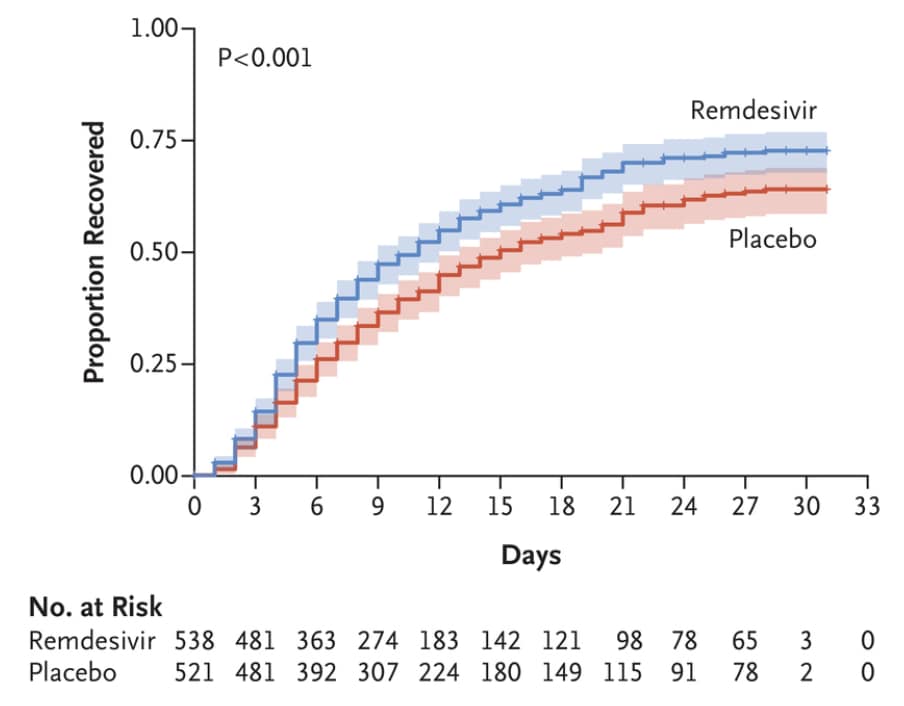
- Among patients treated with remdesivir, recovery time was reduced by 4 days compared with patients in the placebo group (median recovery time: 11 vs. 15 days, p<0.001).
- Patients who received remdesivir were less likely to die than those in the placebo group (14-day mortality rate: 7.1% vs. 11.9%) but this difference was not statistically significant (p=0.70)
Methods: Randomized, double-blind, placebo-controlled trial of hospitalized COVID-19 patients with lower respiratory tract involvement at 60 trial sites, between February 21 and April 19, 2020. Patients received 10 days of intravenous remdesivir (n = 538) or placebo (n = 521) and followed for 29 days. Limitations: Preliminary report.
Figure:
Note: Adapted from Beigel et al. Proportion of all patients who recovered at 29 days after receipt of remdesivir or placebo. From NEJM. DOI: 10.1056/NEJMoa2007764. Copyright © 2020 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
B. Remdesivir for 5 or 10 days in patients with severe Covid-19external icon. Goldman et al. NEJM (May 28, 2020).
Key findings:
- Improved clinical status scores of >2 points (7-point scale) were observed among patients treated with remdesivir for 5 days (64%) or 10 days (54%).
- When controlling for baseline differences, there was no difference in the distribution of clinical status at day 14 of illness between the 2 groups.
Methods: Randomized, open-label, trial of intravenous remdesivir given for 5 or 10 days in 397 hospitalized COVID-19 patients at 55 hospitals, between March 6 and 26, 2020. Clinical status was assessed on a 7-point scale. Limitations: No placebo arm; patients in the 10-day group had worse baseline clinical status than the 5-day group; only 44% in the 10-day treatment group completed a full course of treatment.
Implications of 2 studies (Beigel et al. & Goldman et al.): Remdesivir appears to provide moderate clinical benefit among COVID-19 patients.
Pathogenic viruses such as poliovirus, norovirus, and hepatitis A virus can be spread through microscopic contamination of food, water, or surfaces with the feces from infected people. Detection of viral RNA in feces has raised the question of whether transmission of SARS-CoV-2 can occur via the fecal-oral route.
PEER-REVIEWED
A. SARS-CoV-2 productively infects gut enterocytesexternal icon. Lamers et al. Science (May 1, 2020).
Key findings:
- SARS-CoV-2 is capable of infecting and replicating in a laboratory model of the human small intestine.
- Viral replication occurred at levels lower than in an airway model and decreased after 24 hours (Figure).
- In response to infection with SARS-CoV-2, the intestinal cells produced an antiviral, inflammatory immune response.
Methods: 3-dimensional organoid models of the human small intestine and airway were grown from differentiated adult stem cells. SARS-CoV-2 infection was measured over time by staining and visualizing viral particles and quantifying viral RNA. Limitations: Laboratory study.
Figure
Note: Adapted from Lamer et al. SARS-CoV-2 and SARS-CoV infection of human airway and small intestine models grown from differentiated stem cells. Virus levels (log10 y-axis) were measured over time (x-axis) as the median tissue culture infectious dose (TCID50, y-axis). [h p.i.= hours post infection]. Licensed under CC BY 4.0.
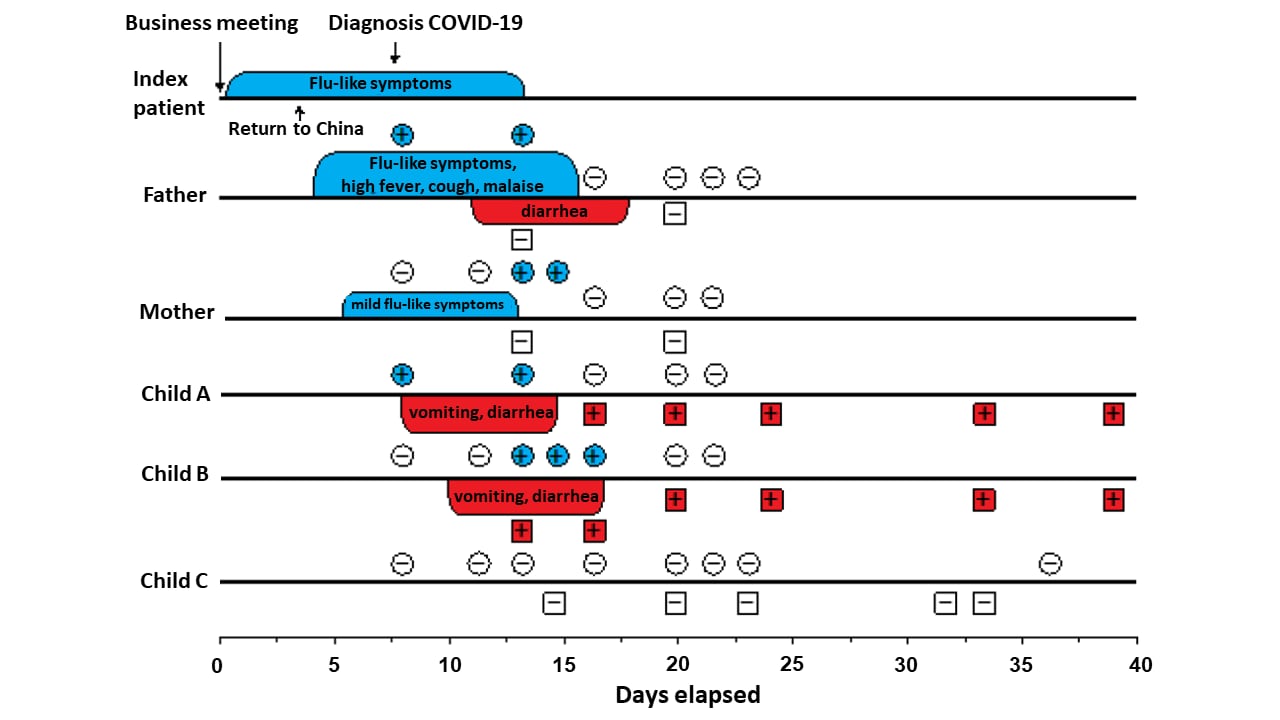
B. Clinical and epidemiological features of a family cluster of symptomatic and asymptomatic SARS-CoV-2 infectionexternal icon. Wolf et al. J Ped Infect Dis (May 22, 2020).
Key findings:
- 4 of 5 family members (80%) had SARS-CoV-2 infection; the uninfected member was a 7-month-old breast-fed infant.
- 2 children, 5 and 2 years old, had gastrointestinal symptoms (vomiting and diarrhea), but no respiratory symptoms (Figure).
- Both children had stool samples RT-PCR positive for SARS-CoV-2 RNA for over 3 weeks after symptoms resolved.
- Neither parent had a positive stool specimen.
Methods: Stool, NP, and OP specimens from all 5 patients were tested for SARS-CoV-2 RNA by RT-PCR. Limitations: Methods for SARS-CoV-2 isolation by culture were described but no results were reported.
Figure:
Note: Adapted from Wolf et al. Timeline of family cluster of SARS-CoV-2 infection with respiratory and gastrointestinal symptoms. PCR-results of respiratory and stool samples. +, positive; –, negative test result. Available via Oxford University Press Public Health Emergency Collection through PubMed Central.
C. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Xiao et al. Emerging Infectious Diseases (May 18, 2020).
Key findings:
- Stool samples from 12 of 28 patients (43%) tested positive for SARS-CoV-2 RNA.
- Live SARS-CoV-2 virus was isolated from the stool of a 78-year old with severe COVID-19 disease who later died.
- For this patient, viral RNA load was consistently higher in stool samples compared with respiratory samples 17 to 28 days after symptoms onset (Figure).
Methods: Stool specimens from 28 patients were tested for SARS-CoV-2 RNA by quantitative RT-PCR. Isolation of SARS-CoV-2 virus using Vero E6 cells (derived from green monkey kidneys) was attempted from 3 of the viral RNA-positive patients. Viral load was assessed for serial stool samples as well NP and OP samples taken from 1 patient during a 10-day period. Limitations: Convenience sample; viral load information available for only 1 patient; no information on the immune status or other clinical history of the patient from whom virus was cultured; detection of viral RNA does not indicate presence of live infectious virus.
Figure:
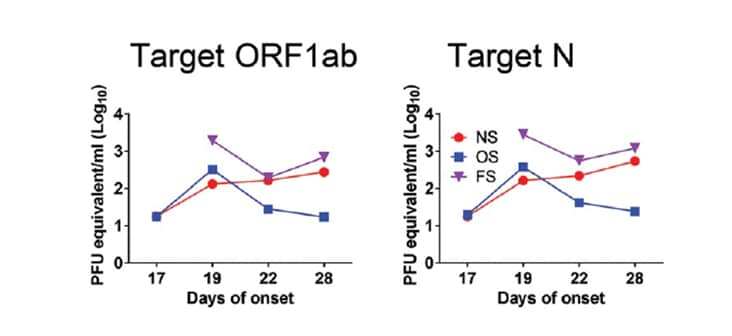
Note: Adapted from Xiao et al. The purple line shows viral RNA load (log10 PFU equivalent/mL on the y-axis) over time in the stool (FS), which remained higher than samples from the nasopharynx (NS) or oropharynx (OS). Two gene regions targeted by quantitative RT-PCR: Target ORF1ab and Target N. Open access journal; all content freely available.
D. Notes from the field: Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the Coronavirus disease 2019 (COVID-19)external icon. Zhang et al. China CDC Weekly (Feb 15, 2020).
Key findings:
- Live SARS-CoV-2 virus was visualized by electron microscopy from a culture inoculated with stool of a person with severe COVID-19 disease 15 days after symptoms onset.
Methods: SARS-CoV-2 virus was visualized from a stool specimen using Vero E6 cells at China CDC laboratory. Limitations: Virus isolated from a single specimen; viral culture methods (e.g., serial passage) inadequately described; medical history and underlying conditions (e.g., immunocompromise) not reported.
Implications of 4 studies (Lamers et al., Wolf et al., Xiao et al., Zhang et al.): SARS-CoV-2 can replicate in human intestinal cells and viral RNA can be detected in stool, often at higher levels and for longer periods of time than in respiratory specimens. However, detection of RNA does not necessarily mean that live, intact virus is present and can cause infection. The cases of live virus isolation described here occur in the backdrop of extensive data from other coronavirus specialty labs that have consistently been unable to isolate live virus from stool samples (e.g. Wölfel et. al., 2020external icon). It is uncertain whether there are factors such as weakened immune function that may lead to shedding of live virus in feces. Based on available evidence, stool appears to pose a very low risk for spread of SARS-CoV-2.
- Frieden et al. Identifying and interrupting superspreading events—implications for control of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases. Discussion of the drivers and prevention of superspreading events of SARS-CoV-2 transmission.
- Oladele et al. The impact of COVID-19 on HIV financing in Nigeria: a call for proactive measuresexternal icon. BMJ Global Health. Describes the impact of COVID-19 on the Nigerian economy and on HIV funding; proposes 6 proactive steps to ensure sustainability of the HIV response.
- Rubin R. Building an army of “disease detectives” to trace COVID-19 contactsexternal icon. JAMA. Discussion of logistics of staffing and conducting contact investigations.
- Giacalone et al. The fear of COVID-19 infection is the main cause of the new diagnoses of hand eczema: report from the frontline in Milanexternal icon. Dermatologic Therapy. Overuse of sanitizers and excessive hand washing leading to eczema.
- Wise J. COVID-19 and thrombosis: what do we know about the risks and treatment?external icon BMJ. Q&A about thrombosis in the context of COVID-19.
- Memtsoudis et al. Obesity as a risk factor for poor outcome in COVID-19-induced lung injury: the potential role of undiagnosed obstructive sleep apnoeaexternal icon. British Journal of Anaesthesia. Discussion of obstructive sleep apnea as a potential contributor to COVID-19 morbidity in obese patients.
- Yu et al. A case of goggle-mask-related impetigo at the time of the COVID-19 pandemicexternal icon. Dermatologic Therapy. Describes impetigo in a health care worker with extended periods of mask use.
- Vital Strategies, World Health Organization. Revealing the Toll of COVID-19: A Technical Package for Rapid Mortality Surveillance and Epidemic Responsepdf iconexternal icon. WHO guidance on implementing rapid mortality surveillance in the context of the COVID-19 pandemic, with a focus on low resource settings.
- Togun et al. Anticipating the impact of the COVID-19 pandemic on TB patients and TB control programmesexternal icon. Annals of Clinical Microbiology & Antimicrobials. Describes effect of COVID-19 on tuberculosis and lessons from experience from tuberculosis control.
- Gefen et al. Pediatric COVID-19-associated rhabdomyolysis: a case reportexternal icon. Pediatric Nephrology. 16-year-old boy is the first pediatric case of severe rhabdomyolysis associated with COVID-19 infection.
Disclaimer: The purpose of the CDC COVID-19 Science Update is to share public health articles with public health agencies and departments for informational and educational purposes. Materials listed in this Science Update are selected to provide awareness of relevant public health literature. A material’s inclusion and the material itself provided here in full or in part, does not necessarily represent the views of the U.S. Department of Health and Human Services or the CDC, nor does it necessarily imply endorsement of methods or findings. While much of the COVID-19 literature is open access or otherwise freely available, it is the responsibility of the third-party user to determine whether any intellectual property rights govern the use of materials in this Science Update prior to use or distribution. Findings are based on research available at the time of this publication and may be subject to change.