Volume 6, Number 6—December 2000
Research
Predominance of HIV-1 Subtype A and D Infections in Uganda
Figure
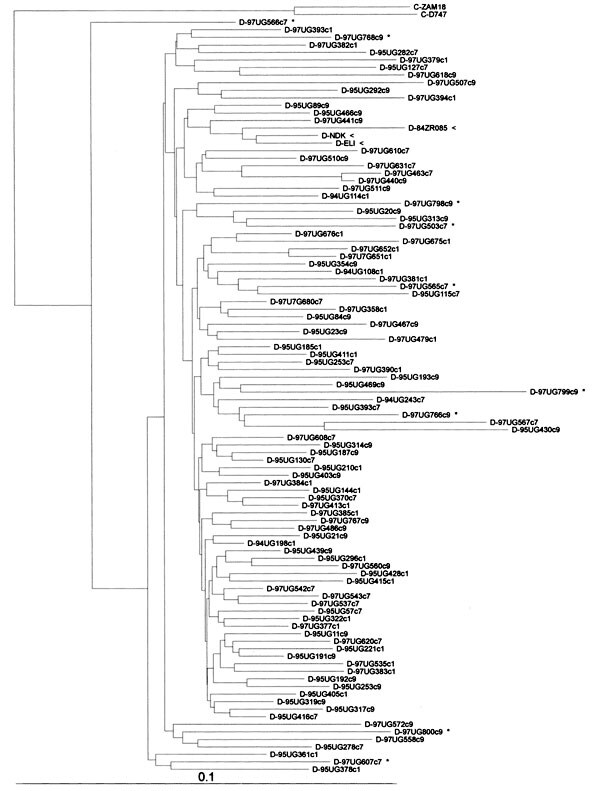
Figure. Phylogenetic classification of env gp41 HIV-1 sequences from Ugandan (UG) patients (GenBank accession numbers for subtypes A and D are pending). Numbers before the abbreviation UG indicate the year of specimen collection; c1, c7, and c9 denote the UVRI, Mulago, and Nsambya clinics, respectively. The trees were constructed on the basis of 354-bp DNA sequences by the neighbor joining method with nucleotide distance datum sets calculated by Kimura's two-parameter approach and rerooted by using SIV-cpz as the outgroup. Arrows indicate reference subtype A and D sequences; asterisks indicate sequences, which decrease the bootstrap value from 90% to 73% in subtype A and from 85% to 55% in subtype D sequences. The scale bar indicates the evolutionary distance of 0.10 nucleotides per position in the sequence. Vertical distances are for clarity only. An automated DNA sequencer (Applied Biosystems Model 373, Foster City, CA) was used to generate sequence data for alignment with the CLUSTAL version V multiple sequence alignment program and subsequent phylogenetic analysis. Phylogenetic relationship of sequences was analyzed by the neighbor joining method (PHYLIP package version 3.5c with and without bootstrapping), and the maximum-likelihood method (fastDNA program, version 1.0.8, which uses randomized data input and global rearrangement). The stability of tree topology was tested by pruning, which consisted of removing one species from the alignment and rerunning the phylogenetic analysis. Accurate subtype determination using env gp41 has been shown to be similar to that based on env C2V3 sequences (22). The gp41 DNA sequences Environment package, and immunodominant regions were analyzed (23). The reference sequences for subtypes A-J, groups O and N, and SIVcpz were retrieved from the 1997 HIV-1 Molecular Immunology Database (Los Alamos National Laboratory, Los Alamos, NM).
References
- Hu DJ, Dondero TJ, Rayfield MA, George JR, Schochetman G, Jaffe HW, The emerging genetic diversity of HIV: The importance of global surveillance for diagnostics, research, and prevention. JAMA. 1996;275:210–6. DOIPubMedGoogle Scholar
- Ou CY, Takebe Y, Luo CC, Kalish ML, Auwanit W, Bandea C, Wide distribution of two subtypes of HIV-1 in Thailand. AIDS Res Hum Retroviruses. 1992;8:1471–2.PubMedGoogle Scholar
- Weniger BG, Limpakarnjanarat K, Ungchusak K, Thanprasertsuk S, Choopanya K, Vanichseni S, The epidemiology of HIV infection and AIDS in Thailand. AIDS. 1991;5:S71–85. DOIPubMedGoogle Scholar
- McCutchan FE, Hegerich PA, Brennan TP, Phanuphak P, Singharaj P, Jugsudee A, Genetic variants of HIV-1 in Thailand. AIDS Res Hum Retroviruses. 1992;8:1887–95. DOIPubMedGoogle Scholar
- Wright NH, Vanichseni S, Akarasewi P, Wasi C, Choopanya K. Was the 1988 HIV epidemic among Bangkok's injecting drugs users a common source outbreak? AIDS. 1994;8:529–32. DOIPubMedGoogle Scholar
- Kalish ML, Baldwin A, Raktham S, Wasi C, Luo CC, Schochetman G, The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials. AIDS. 1995;9:851–7. DOIPubMedGoogle Scholar
- Wasi C, Herring B, Raktham S, Vanichseni S, Mastro TD, Young NL, Determination of HIV-1 subtypes in injecting drug users in Bangkok, Thailand, using peptide-binding enzyme immunoassay and heteroduple mobility assay: evidence of increasing infection with HIV-1 subtype E. AIDS. 1995;9:843–9. DOIPubMedGoogle Scholar
- Subbarao S, Limpakarnjanarat K, Mastro TD, Bhumisawasdi J, Warachit P, Jayavasu C, HIV-1 in Thailand, 1994-1995: persistence of two subtypes with low genetic diversity. AIDS Res Hum Retroviruses. 1998;14:319–27. DOIPubMedGoogle Scholar
- Janssens W, Buve A, Nkengasong JN. The puzzle of HIV-1 subtypes in Africa. AIDS. 1997;11:705–12. DOIPubMedGoogle Scholar
- Barin F, Courouce AM, Pillonel J, Buzelay L; Retrovirus Study Group of the French Society of Blood Transfusion. Increasing diversity of HIV-1M serotypes in French blood donors over a 10-year period (1985-1995). AIDS. 1997;11:1503–8. DOIPubMedGoogle Scholar
- M üller-Trutwin MC, Chaix ML, Letourneur F, Begaud E, Beaumont D, Deslandres A, Increase of HIV-1 subtype A in Central African Republic. J Acquir Immune Defic Syndr. 1999;21:164–1.PubMedGoogle Scholar
- Mastro TD, Kunanusont C, Dondero TJ, Wasi C. Why do HIV-1 subtypes segregate among persons with different risk behaviors in South Africa and Thailand? AIDS. 1997;11:113–6. DOIPubMedGoogle Scholar
- Oram JD, Downing RG, Roff M, Serwankambo N, Clegg JCS, Featherstone AS, Sequence analysis of the V3 loop regions of the env genes of Ugandan human immunodeficiency proviruses. AIDS Res Hum Retroviruses. 1991;1:605–14. DOIPubMedGoogle Scholar
- Albert J, Franzen L, Jansson M, Scarlatti G, Kataaha PK, Katabira E, Ugandan HIV-1 V3 loop sequences closely related to the U.S./European consensus. Virology. 1992;190:674–81. DOIPubMedGoogle Scholar
- WHO Network for HIV Isolation and Characterization. HIV type 1 variation in World Health Organization-sponsored vaccine evaluation sites: genetic screening, sequence analysis, and preliminary biological characterization of selected viral strains. AIDS Res Hum Retroviruses. 1994;10:1327–43. DOIPubMedGoogle Scholar
- Bruce C, Clegg C, Featherstone A, Smith J, Biryahawaho B, Downing R, Presence of multiple genetic subtypes of human immunodeficiency virus type 1 proviruses in Uganda. AIDS Res Hum Retroviruses. 1994;10:1543–50. DOIPubMedGoogle Scholar
- Smith JD, Bruce CB, Featherstone AS, Downing RG, Biryahawaho B, Clegg , . Reactions of Ugandan antisera with peptides encoded by V3 loop epitopes of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:577–83. DOIPubMedGoogle Scholar
- Brennan CA, Lund JK, Golden A, Yamaguchi J, Vallari AS, Phillips JF, Serologic and phylogenetic characterization of HIV-1 subtypes in Uganda. AIDS. 1997;11:1823–32. DOIPubMedGoogle Scholar
- Rayfield MA, Downing RG, Baggs J, Hu DJ, Pieniazek D, Luo CC, A molecular epidemiologic survey of HIV in Uganda. AIDS. 1998;12:521–7. DOIPubMedGoogle Scholar
- Luo CC, Downing RG, dela Torre N, Baggs J, Hu DJ, Respess RA, The development and evaluation of a probe hybridization method for subtyping HIV type 1 infection in Uganda. AIDS Res Hum Retroviruses. 1998;14:691–4. DOIPubMedGoogle Scholar
- Yang C, Pieniazek D, Owen SM, Fridlund C, Nkengasong J, Mastro TD, Detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma by using highly sensitive and specific generic primers. J Clin Microbiol. 1999;37:2581–6.PubMedGoogle Scholar
- Pieniazek D, Yang C, Lal RB. Phylogenetic analysis of gp41 envelope of HIV-1 groups M, N, and O provides an alternate region for subtype determination. In: Korber B, Foley B, McCutchan F, Mellors JW, Hahn BH, Sodroski J, et al, editors. Human retroviruses and AIDS 1998. Los Alamos: Los Alamos National Laboratory;1998:III-112-17.
- Dorn J, Masciotra S, Yang C, Downing R, Biryahwaho B, Mastro TD, Analysis of genetic variability within the immunodominant epitopes of envelope gp41 from HIV-1 Group M and its impact on HIV-1 antibody detection. J Clin Microbiol. 2000;38:773–80.PubMedGoogle Scholar
- Downing R, Pieniazek D, Hu DJ, Biryahawaho B, Fridlund C, Rayfield MA, Genetic characterization and phylogenetic analysis of HIV-1 subtype C from Uganda. AIDS Res Hum Retroviruses. 2000;16:815–9. DOIPubMedGoogle Scholar
- Hu DJ, Buvé A, Baggs J, van der Groen G, Dondero TJ. What role does HIV-1 subtype play in transmission and pathogenesis? An epidemiological perspective. AIDS. 1999;3:873–81. DOIPubMedGoogle Scholar
- Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature. 1995;374:124–6. DOIPubMedGoogle Scholar