Volume 30, Number 4—April 2024
Research
Bus Riding as Amplification Mechanism for SARS-CoV-2 Transmission, Germany, 20211
Figure 3
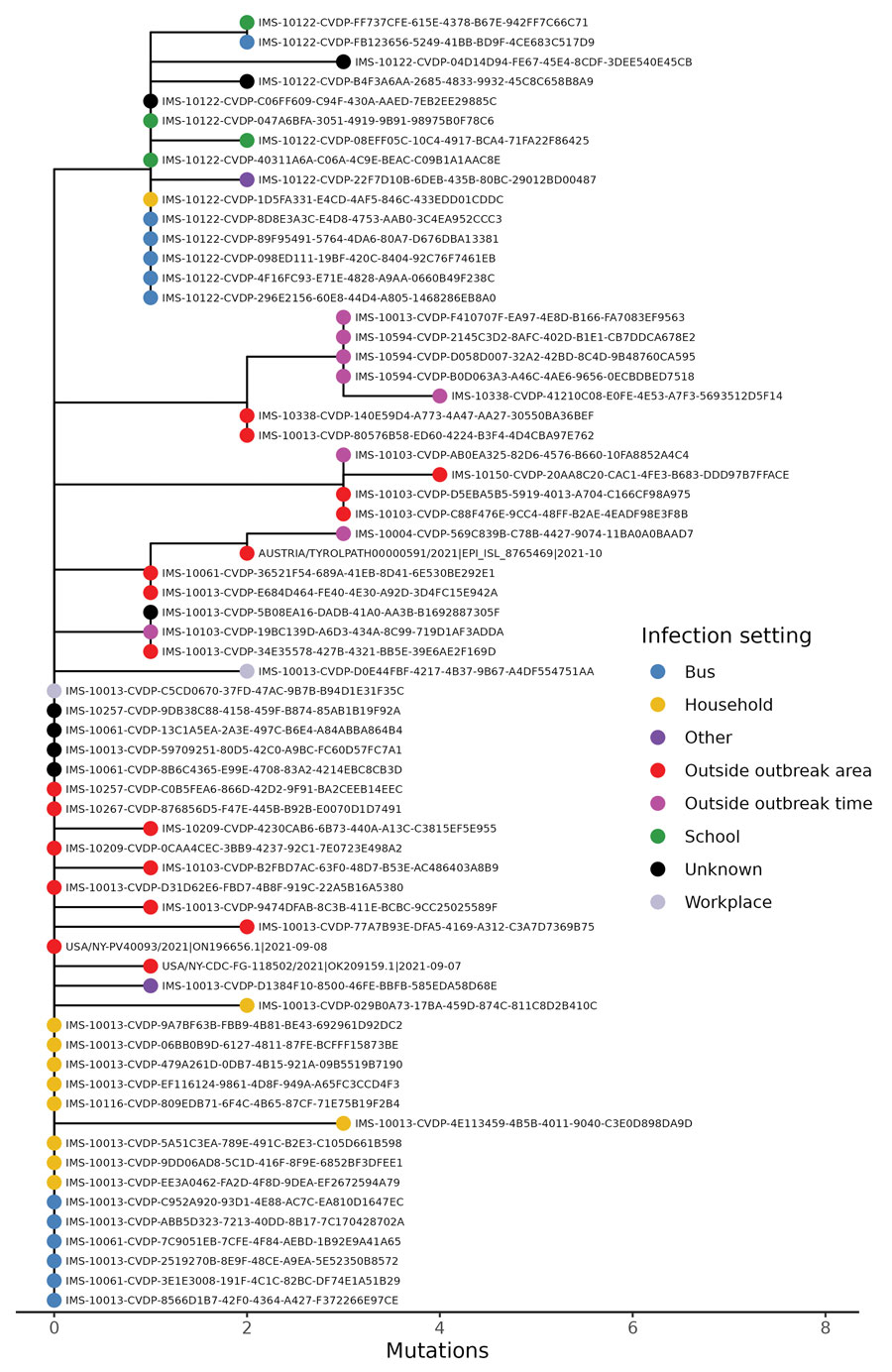
Figure 3. Sequences from samples from case-patients in COVID-19 outbreak, Hesse, Germany, 2021 (red circles). Sequences cluster tightly together when placed in a global phylogenetic tree, with the exception of 1 outlier. The outlier sequence from 1 air traveler was removed from the outbreak case-patients on the basis of lack of sequence similarity compared with other outbreak sequences. The global phylogenetic tree includes all SARS-CoV-2 sequences GISAID (https://gisaid.org), GenBank, COVID-19 Genomics UK Consortium (https://www.cogconsortium.uk), and the China National Center for Bioinformation (https://www.cncb.ac.cn) databases as of July 16, 2023 (total ≈15 million sequences) and was downsampled to ≈2,000 sequences for easier visualization.
1Preliminary results from this study were presented at the ESCAIDE conference; November 24, 2022; Stockholm, Sweden.
2These first authors contributed equally to this article.
3These senior authors contributed equally to this article.