Volume 30, Number 4—April 2024
Research Letter
Case of Human Orthohantavirus Infection, Michigan, USA, 2021
Abstract
Orthohantaviruses cause hantavirus cardiopulmonary syndrome; most cases occur in the southwest region of the United States. We discuss a clinical case of orthohantavirus infection in a 65-year-old woman in Michigan and the phylogeographic link of partial viral fragments from the patient and rodents captured near the presumed site of infection.
Orthohantaviruses are negative-sense, enveloped RNA viruses that are transmitted by host reservoirs, such as rodents, to humans. Human infection occurs through inhalation of aerosolized viral particles from host excreta, such as urine or feces, often in enclosed spaces during infestations. New World orthohantavirus infection results in hantavirus cardiopulmonary syndrome (HCPS), which consists of febrile illness with edema and respiratory failure (1). In the United States, most HCPS cases occur in the Southwest and have a ≈35% mortality rate (2).
The dominant orthohantavirus that causes HCPS in the United States is Sin Nombre virus (SNV), which is thought to be carried and transmitted by the western deer mouse (Peromyscus sonoriensis). New York virus (NYV) is another pathogenic variant of orthohantavirus that is found in white-footed deer mice (Peromyscus leucopus); cases occur primarily in the Northeast region of the country (3). Although multiple host reservoirs for orthohantaviruses are distributed throughout the United States, most human cases are caused by SNV (4,5).
In early May 2021, a previously healthy 65-year-old woman visited an emergency department in Washtenaw County, Michigan, USA, with febrile prodrome of 3–6 days, thrombocytopenia, mild transaminase elevation, and acute hypoxic respiratory failure of unclear etiology requiring intubation. An extensive infectious disease workup was conducted, and physicians initially ruled out such pulmonary pathogens as SARS-CoV-2, common respiratory viruses, fungal agents, and Legionella spp. The family was interviewed to obtain a travel and animal exposure history, which revealed that the patient had not traveled outside of Michigan in the previous year. The interview also confirmed that the patient had not consumed unpasteurized dairy or undercooked meat, had a mostly indoor dog, lived near a natural area but used trails/sidewalks, and had no known rodent infestation in the home. However, the spouse reported that the patient had spent time recently cleaning out a relative’s home that had been uninhabited for 2 years and was infested with mice.
Results of a tickborne disease panel were negative, but hantavirus antibody testing performed at a commercial lab showed positive results for both IgM and IgG. The treating hospital notified the Michigan Department of Health and Human Services of a case of HCPS. Confirmatory hantavirus testing was arranged and confirmed with the Centers for Disease Control and Prevention, using serum samples collected from hospitalization.
Trapping was performed in and around the suspected site of exposure (relative’s home) using Sherman folding traps (https://shermantraps.com; 94 trap nights), resulting in 12 rodents captured (12.8% trap success) under an approved animal-use protocol (6). Trapping was conducted 12 days after the patient was released from the hospital. Researchers observed signs of previous trapping efforts; 5 unusable Peromyscus mouse carcasses were found in snap traps in the residential basement. Signs of infestation were evident. Of the 12 trapped rodents, 3 (25%) were P. leucopus mice, 1 (8%) was a Northern short-tailed shrew (Blarina brevicauda), and 8 (67%) were Eastern chipmunks (Tamias striatus) (Table). The surrounding flora consisted of lawns, shrubs, and an evergreen windbreak near a public trail.
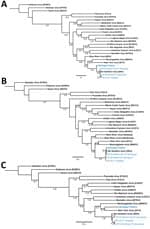
Using quantitative reverse transcription PCR, we screened lung, liver, brown fat, or kidney tissue from captured rodents and from a plasma sample of the patient obtained during hospitalization (6). Brown fat and kidney tissue from 2 P. leucopus mice and lung tissue from 2 T. striatus chipmunks tested positive for SNV. Three fragments were obtained from the patient sample, 1 for the short segment (480 bp), 1 for the medium segment (283 bp), and 1 for the large segment (377 bp). Similar fragments were also generated from 3 of the 4 infected rodents; all sequences are publicly available in GenBank (accession nos. OR428177–88). We compared fragments by using phylogenetic analysis against several known orthohantavirus reference sequences to determine potential identification. The partial sequences of SNV short and medium segments from the patient formed a phylogenetic lineage with SNV sequences from the rodents collected in or near the suspected site of exposure in Michigan. However, the patient’s large fragment formed a lineage with NYV, suggesting that this species may be an SNV or NYV variant (Figure).
Previously, we identified the likely site of rodent-to-human SNV transmission in a patient case study (6). Here, we attempted a similar approach but were only able to generate partial sequences for the patient sample, which we compared with captured rodents. Orthohantavirus incubation periods can be up to several weeks after exposure (7), which may impact the timeliness of trapping efforts. We found infected P. leucopus mice and T. striatus chipmunks at the site of exposure, both of which have been reported to carry NYV or SNV; P. leucopus mice are susceptible and capable vessels for SNV replication after laboratory infection (6,8–10). This finding suggests that orthohantaviruses may not be as species host–restricted as previously thought. Further studies are warranted to clarify (or define) orthohantavirus species in Michigan to anticipate the risk for patient infection. Increasing surveillance and diagnostic efforts can enable prospective detection of circulating viruses.
Dr. Goodfellow is a recent graduate from the University of New Mexico Health Sciences Center. His primary research interests are emerging and re-emerging infectious diseases, surveillance efforts, and science policy.
Acknowledgments
We thank Michigan’s Department of Health and Human Services and Washtenaw County Health Department for collaboration, along with the Centers for Disease Control and Prevention Epidemiology and Ecology teams that participated in this response.
This work was supported in part by a University of New Mexico School of Medicine Research Allocation Committee (UNM SOM RAC) grant (S.B.B.) and an NIH/NIAID grant (AI-I7-042 U19, K.C.). S.M.G. was supported by UNM HSC Infectious Disease and Inflammation Program NIH grant T32AI007.
References
- Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–7. DOIPubMedGoogle Scholar
- Akram SM, Mangat R, Huang B. Hantavirus cardiopulmonary syndrome. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024.
- Hjelle B, Lee SW, Song W, Torrez-Martinez N, Song JW, Yanagihara R, et al. Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: genetic characterization of the M genome of New York virus. J Virol. 1995;69:8137–41. DOIPubMedGoogle Scholar
- Rollin PE, Ksiazek TG, Elliott LH, Ravkov EV, Martin ML, Morzunov S, et al. Isolation of black creek canal virus, a new hantavirus from Sigmodon hispidus in Florida. J Med Virol. 1995;46:35–9. DOIPubMedGoogle Scholar
- Ksiazek TG, Nichol ST, Mills JN, Groves MG, Wozniak A, McAdams S, et al. Isolation, genetic diversity, and geographic distribution of Bayou virus (Bunyaviridae: hantavirus). Am J Trop Med Hyg. 1997;57:445–8. DOIPubMedGoogle Scholar
- Goodfellow SM, Nofchissey RA, Schwalm KC, Cook JA, Dunnum JL, Guo Y, et al. Tracing transmission of Sin Nombre virus and discovery of infection in multiple rodent species. J Virol. 2021;95:
e0153421 . DOIPubMedGoogle Scholar - Vial PA, Valdivieso F, Mertz G, Castillo C, Belmar E, Delgado I, et al. Incubation period of hantavirus cardiopulmonary syndrome. Emerg Infect Dis. 2006;12:1271–3. DOIPubMedGoogle Scholar
- Mills JN, Ksiazek TG, Ellis BA, Rollin PE, Nichol ST, Yates TL, et al. Patterns of association with host and habitat: antibody reactive with Sin Nombre virus in small mammals in the major biotic communities of the southwestern United States. Am J Trop Med Hyg. 1997;56:273–84. DOIPubMedGoogle Scholar
- Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, et al. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis. 1994;169:1271–80. DOIPubMedGoogle Scholar
- Quizon K, Holloway K, Iranpour M, Warner BM, Deschambault Y, Soule G, et al. Experimental infection of peromyscus species rodents with Sin Nombre virus. Emerg Infect Dis. 2022;28:1882–5. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: March 11, 2024
Table of Contents – Volume 30, Number 4—April 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Steven B. Bradfute, University of New Mexico, 915 Camino de Salud NE, 3190 IDTC Bldg 0289, Albuquerque, NM 87131, USA
Top