Volume 29, Number 9—September 2023
Dispatch
Population Analysis of Escherichia coli Sequence Type 361 and Reduced Cefiderocol Susceptibility, France
Abstract
Cefiderocol resistance is increasingly reported in New Delhi metallo-β-lactamase–producing Enterobacterales. Genomic and phenotypic analysis of Escherichia coli sequence type 361, a primary clone causing carbapenemase spread in France, revealed mutations leading to cefiderocol resistance. Continued genomic surveillance of carbapenem-resistant Enterobacterales could clarify prevalence of cefiderocol-resistant E. coli in Europe.
Few last-line antimicrobial agents effectively treat infections caused by New Delhi metallo-β-lactamase (NDM)–producing Enterobacterales (1). Cefiderocol is a novel synthetic conjugate siderophore cephalosporin that is more stable against β-lactamase hydrolysis than classical cephalosporins (2). However, several acquired cefiderocol-resistance mechanisms have been described in Enterobacterales, including increased blaNDM copy numbers (3), specific blaKPC variants (4), structural change in AmpC (5), and mutations or inactivation of siderophore receptors (6). Specific polymorphisms in penicillin-binding protein 3 (PBP3), the target of cefiderocol, also have been reported in Acinetobacter and Escherichia coli (7–9). However, prevalence of those polymorphisms and effects of cumulative resistance mechanisms have not been fully evaluated.
Since 2012, the French National Reference Center (F-NRC) for Antimicrobial Resistance has conducted active nationwide surveillance of carbapenemase-producing Enterobacterales (CPE). In 2022, the percentage of E. coli sequence type (ST) 361 isolates sent to F-NRC doubled to 1.2% from 0.6% of CPE in 2021. We characterized emerging E. coli ST361 in France and investigated cefiderocol resistance among CPE.
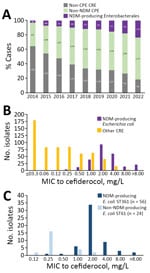
Since 2014, prevalence of NDM-producing Enterobacterales has been increasing in France (Figure 1, panel A). Among NDM producers, we observed a polyclonal dissemination of E. coli isolates, but 50% of isolates were from 4 main clones (ST410, ST167, ST361, and ST405), as reported in other countries in Europe (Appendix 1 Figure 1) (10). E. coli ST410, ST167, and ST405 have been characterized at the genomic level (3,8,11), but ST361 characteristics remain unclear.
During July 1, 2021–June 30, 2022, we investigated all (n = 856) nonduplicate carbapenem-nonsusceptible E. coli isolates sent to F-NRC. We used Sensititer broth microdilution (ThermoFisher, https://www.thermofisher.com), as previously described (12), to measure MICs of aztreonam, ceftazidime-avibactam, imipenem, meropenem, and cefiderocol (Figure 2). Of note, the Mueller–Hinton broths used were from batches not affected by the manufacturer’s withdrawal relayed by European Committee on Antimicrobial Susceptibility Testing (https://www.eucast.org/ast-of-bacteria/warnings). Among tested isolates, 774 were CPE, including 243 NDM producers. The MIC50 (MIC to inhibit growth of 50% of isolates) of cefiderocol was higher (2 mg/L) for NDM producers among isolates tested compared with other carbapenem-resistant E. coli (0.12 mg/L) (Figure 1, panel B), as previously reported (12).
To genomically characterize E. coli ST361, we added all (n = 51) ST361 isolates sent to F-NRC during 2015–2021 to the 29 isolates collected during the study period. We conducted short-read sequencing on those 80 isolates by using the NextSeq500 system (Illumina, https://www.illumina.com). We assembled sequences by using Shovill 1.1.0 (https://github.com/tseemann/shovill) and SPAdes 3.14.0 (https://github.com/ablab/spades) under GenBank BioProject no. PRJNA925451 (Appendix 2 Table 1). We used Resfinder 4.1 (13) to analyze resistome content and PlasmidFinder 2.1 (14) to analyze replicon content (Appendix 2 Table 2).
Among 80 E. coli ST361 isolates, 50 produced NDM carbapenemase, 49 of which were NDM-5; another 20 produced oxacillinase 48–like carbapenemase; 6 coproduced NDM-5 with another carbapenemase; and 4 did not produce carbapenemase (Figure 2). Analysis of cefiderocol MIC distribution for ST361 showed that isolates with MICs >2 produced NDM, but that analysis also suggested that mechanisms besides NDM are involved in cefiderocol resistance (Figure 1, panel C). Thus, we analyzed the blaNDM gene copy number on CLC Genomics Workbench 21.0 (QIAGEN, https://www.qiagen.com), where we mapped the raw data (fastq reads) on the genome (fasta) of each corresponding E. coli sequence. Then we normalized the average coverage of blaNDM mapping reads to the average coverage of 10 different chromosomal genes used as references. However, correlation analysis did not reveal an association between blaNDM gene number and the cefiderocol MIC (data not shown). Then we used E. coli K-12 MG1655 (GenBank accession no. NC_000913) as a reference to investigate cirA, fiu, fepA, fepB, fecA, fhuA, tonB, pcnB, exbB, exbD, baeS/baeR, and ompR/envZ gene mutations involved in siderophore-iron uptake. To eliminate polymorphisms linked to the ST itself, we only considered amino acid substitutions not shared by all ST361 isolates. A total of 14 (18%) isolates displayed a mutation in 1 of those genes (Appendix 2 Table 1). Overall, analysis of variance multiple parameter correlation analysis in RStudio 2022.07.1 (The R Foundation for Statistical Computing, https://www.r-project.org) revealed that blaNDM (p = 0.0035) or chromosomal mutations (p = 0.0033) within a siderophore receptor were associated with higher cefiderocol MICs.
We also analyzed the preferential cefiderocol target, PBP3. That analysis revealed that compared with the reference, 76 isolates shared a common allele that had a 4 amino acid insertion (YRIN motif) at position 333 and 3 substitutions (Q227H, E349K, and I532L). Two isolates had a different allele with a YRIK insertion and an A412V substitution, and 2 isolates had no insertions or mutations. Of note, the 3 different alleles were associated with 3 different nodes on the phylogenetic tree, indicating an evolution process that probably involved chromosomal recombination (Figure 2), as described for ST410 (11). The YRIN(K) motif insertion has been described to be involved in cephalosporin and aztreonam resistance (8,9,11). To study the effect of the YRIN(K) motif insertion on cefiderocol resistance, we performed susceptibility testing on the reference strain and its isogenic PBP3 encoding gene mutant with YRIN insertion (11). We transformed both strains by plasmid topoisomerase-based cloning blaNDM-1 to increase the basal range of cefiderocol MIC concentrations in the microbroth dilution technique. The YRIN insertion resulted in a 4-fold increase in cefiderocol MIC, from <0.03 mg/L to 0.125 mg/L in the YRIN ftsI chromosomal mutant.
We also analyzed all (n = 321) available ST361 genomes and metadata in EnteroBase (University of Warwick, https://warwick.ac.uk/fac/sci/med/research/biomedical/mi/enterobase) on October 1, 2022 (Appendix 1 Figures 3, 4; Appendix 2 Table 3). The 401-isolate phylogenetic tree showed that isolates from F-NRC were distributed within several main branches (Appendix 1 Figure 3), confirming our collection’s diversity. Of note, the YRIN(K) insertion occurred in only 36% of the EnteroBase genomes but occurred in 97% of NDM producers; however, only 7% of non–NDM producers had the modified alleles. The phylogenetic tree enabled visualization of this strong association between occurrence of NDM and PBP3 alleles possessing the YRIN(K) insertion. Furthermore, specifying isolate locations revealed international ST361 circulation.
We also examined genomes sequenced at F-NRC during 2015–2022 that are from 3 other predominant STs disseminating NDM-5 in France. Among those genomes, we noted a high prevalence of YRIN(K) insertion in PBP3, namely in 98% of ST410 (n = 273), 92% of ST167 (n = 184), and 86% of ST405 (n = 122), regardless of β-lactamase content (Appendix 1 Figure 5, panel A). YRIN(K) insertion prevalence was only 4% in E. coli ST131 (n = 166), another high-risk clone associated with multiple β-lactamases (15). Distribution analysis of cefiderocol MICs in ST410, ST167, and ST405, excluding NDM-producing isolates, revealed a MIC50 of 1 mg/L, confirming the role of the genetic background in reduced cefiderocol susceptibility (Appendix 1 Figure 5, panel B).
Our results highlight the emergence of NDM-producing E. coli ST361 associated with reduced cefiderocol susceptibility in France. Emergence resulted from a combination of factors: modified PBP3, a strong association with NDM-5 carbapenemase, and frequent chromosomal mutations in genes involved in siderophore-iron uptake. No feature alone is sufficient to confer cefiderocol resistance, according to published clinical breakpoints (https://www.eucast.org/clinical_breakpoints), but the combined mechanisms appear to confer resistance.
In conclusion, our study revealed that E. coli ST361 is becoming a key player in NDM-5 carbapenemase dissemination, and its genetic background confers reduced cefiderocol susceptibility. E. coli ST361 has only been sporadically reported, but its prevalence might be underestimated. To further assess prevalence and spread of cefiderocol-resistant E. coli in Europe, each country should continue nationwide genomic surveillance of carbapenemase-resistant bacteria.
Dr. Jousset is an assistant professor at the Institut National de la Santé et de la Recherche Médicale (INSERM), Paris, France. Her main research interests include epidemiology, genetics, and biochemistry of β-lactamases in gram-negative bacteria.
Acknowledgment
This work was supported by grants from the French National Research Agency, project Seq2Diag PPR Antibioresistance (grant no. ANR-20-PAMR-0010).
References
- Wright H, Bonomo RA, Paterson DL. New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect. 2017;23:704–12. DOIPubMedGoogle Scholar
- Wang C, Yang D, Wang Y, Ni W. Cefiderocol for the treatment of multidrug-resistant gram-negative bacteria: a systematic review of currently available evidence. Front Pharmacol. 2022;13:
896971 . DOIPubMedGoogle Scholar - Simner PJ, Mostafa HH, Bergman Y, Ante M, Tekle T, Adebayo A, et al. Progressive development of cefiderocol resistance in Escherichia coli during therapy is associated with an increase in blaNDM-5 copy number and gene expression. Clin Infect Dis. 2022;75:47–54. DOIPubMedGoogle Scholar
- Hobson CA, Cointe A, Jacquier H, Choudhury A, Magnan M, Courroux C, et al. Cross-resistance to cefiderocol and ceftazidime-avibactam in KPC β-lactamase mutants and the inoculum effect. Clin Microbiol Infect. 2021;27:1172.e7–10. DOIPubMedGoogle Scholar
- Shields RK, Iovleva A, Kline EG, Kawai A, McElheny CL, Doi Y. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin Infect Dis. 2020;71:2713–6. DOIPubMedGoogle Scholar
- Klein S, Boutin S, Kocer K, Fiedler MO, Störzinger D, Weigand MA, et al. Rapid development of cefiderocol resistance in carbapenem-resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin Infect Dis. 2022;74:905–8. DOIPubMedGoogle Scholar
- Malik S, Kaminski M, Landman D, Quale J. Cefiderocol resistance in Acinetobacter baumannii: roles of β-lactamases, siderophore receptors, and penicillin binding protein 3. Antimicrob Agents Chemother. 2020;64:e01221–20. DOIPubMedGoogle Scholar
- Wang Q, Jin L, Sun S, Yin Y, Wang R, Chen F, et al. Occurrence of high levels of cefiderocol resistance in carbapenem-resistant Escherichia coli before its approval in China: a report from China CRE-Network. Microbiol Spectr. 2022;10:
e0267021 . DOIPubMedGoogle Scholar - Sato T, Ito A, Ishioka Y, Matsumoto S, Rokushima M, Kazmierczak KM, et al. Escherichia coli strains possessing a four amino acid YRIN insertion in PBP3 identified as part of the SIDERO-WT-2014 surveillance study. JAC Antimicrob Resist. 2020;2:dlaa081.
- Chakraborty T, Sadek M, Yao Y, Imirzalioglu C, Stephan R, Poirel L, et al. Cross-border emergence of Escherichia coli producing the carbapenemase NDM-5 in Switzerland and Germany. J Clin Microbiol. 2021;59:e02238–20. DOIPubMedGoogle Scholar
- Patiño-Navarrete R, Rosinski-Chupin I, Cabanel N, Gauthier L, Takissian J, Madec JY, et al. Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med. 2020;12:10. DOIPubMedGoogle Scholar
- Bonnin RA, Emeraud C, Jousset AB, Naas T, Dortet L. Comparison of disk diffusion, MIC test strip and broth microdilution methods for cefiderocol susceptibility testing on carbapenem-resistant enterobacterales. Clin Microbiol Infect. 2022;28:1156.e1–5. DOIPubMedGoogle Scholar
- Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491–500. DOIPubMedGoogle Scholar
- Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. DOIPubMedGoogle Scholar
- Dautzenberg MJD, Haverkate MR, Bonten MJM, Bootsma MCJ. Epidemic potential of Escherichia coli ST131 and Klebsiella pneumoniae ST258: a systematic review and meta-analysis. BMJ Open. 2016;6:
e009971 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: August 08, 2023
Table of Contents – Volume 29, Number 9—September 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Agnès B. Jousset, Service de Bactériologie-Hygiène, Hôpital de Bicêtre, 78 rue du Général Leclerc, 94275 Le Kremlin-Bicêtre CEDEX, France
Top