Volume 28, Number 6—June 2022
Research Letter
Molecular Diagnosis of Pseudoterranova decipiens Sensu Stricto Infections, South Korea, 2002‒2020
Figure
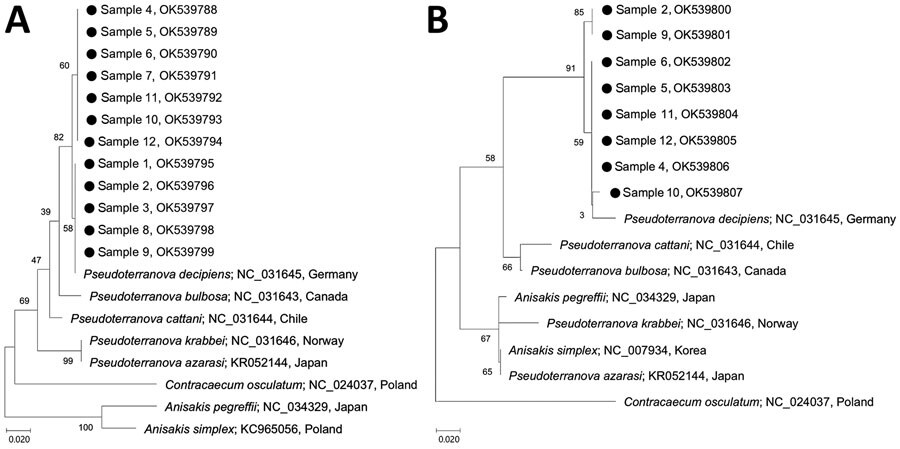
Figure. Phylogenetic analyses of Pseudoterranova nematode larvae extracted from 12 health check-up patients in South Korea, 2002–2020 (black dots), in comparison with other anisakid species. A) mitochondrial cytochrome oxidase c gene sequences; B) mitochondrial NADH dehydrogenase gene sequences. Trees were constructed by using the neighbor-joining method based on the Kimura 2-parameter model of nucleotide substitution with 1,000 bootstrap replications and viewed by using MEGA-X (https://www.megasoftware.net). GenBank accession numbers and country of origin are provided for reference sequences. Details of patient information for the 12 samples from this study are provided in Appendix Table 1). Numbers along branches are bootstrap values. Scale bars indicate nucleotide substitutions/site.
1These authors contributed equally to this article.