Identification of filamentous fungi using MALDI-ToF using the Bruker Biotyper
- 1.0 Purpose
- 2.0 Definitions
- 3.0 Equipment: Properly maintained and calibrated per institutions protocols
- 4.0 Reagents/Supplies/Media
- 5.0 Safety: All institutional safety procedures must be followed in the performance of this standard operating procedure.
- 6.0 Sample Preparation Information / Processing/ Media
- 7.0 Reagent Preparation
- 8.0 Extraction Procedure
3.1
Biological Safety Cabinet
3.2
MALDI-ToF MS Bruker Biotyper
4.1
100 % Ethanol-Sterile Solution: example, SIGMA (Catalog # 459836), or equivalent. Always use Ethanol labeled “molecular biology reagent”.
4.2
Sterile WFI Quality Cell Culture Grade Water: example, Fisher Scientific (Product # L4321849), or equivalent.
4.3
Sabouraud’s Dextrose Agar (Emmons): Plates and slants; example, Fisher Scientific (Product #L21827), or equivalent.
4.4
Reusable polished steel MALDI target plate; 96 sample positions (Bruker Product # 8280800)
4.5
Bruker matrix HCCA (HCCA = α-Cyano-4-hydroxycinnamic acid): (Product # 255344) Prepare by adding 250 µl of Bruker Standard Solvent, vortex until liquid is clear.
4.6
70% Formic Acid: example SIGMA (Catalogue # 27001), diluted using sterile WFI Quality cell culture grade water
4.7
Bacterial test standard (BTS): Bruker (Product # 255343)
4.7.1
Prepare BTS according to manufacturer’s instructions and store at ≤ -18C until ready for use.
4.8
Bruker Standard Solvent (Sigma Aldrich #900666)
4.9
SWUBE Applicator; example VWR (Catalog # 90001-126), or equivalent
4.10
1.5 mL Microcentrifuge tubes; example Fisher Scientific (Product# 05-408-129), or equivalent
4.11
1.0 mm Zirconia/Silica Beads; example Fisher Scientific (Catalog # NC9847287), or equivalent
5.1
Standard personal protective equipment should follow institutional guidelines but should consist of at least gowns, gloves, and safety glasses which should be worn at all times.
5.2
All set-up procedures should be performed in a biological safety cabinet.
5.3
Chemical Safety
5.3.1
Formic Acid: Flammable, Toxic and Corrosive
5.3.2
α-Cyano-4-hydroxycinnamic acid (HCCA): Skin irritant, Serious eye irritant.
6.1
Growth
6.1.1
Plates incubated for 3-7 days should be used for routine microorganism identification; slow-growing molds (e.g. Rhinocladiella) may need to grow for 7-10 days before testing.
6.2
Storage
6.2.1
Storing plates at room temperature for 0-5 days is acceptable; growth temperature (25°C vs 37°C) have little effect on results.
NOTE: Do Not use organisms that have been stored at 4°C or lower as this has a negative impact on quality of spectra and reproducibility.
7.1
For each run, include Aspergillus fumigatus ATCC MYA-3626 and Aspergillus flavus ATCC MYA-3631 (or other validated molds) as positive controls
8.1
Extraction Procedure
NOTE: Perform following steps in a biological safety cabinet to ensure safety and sterility.
8.1.1
Perform extraction procedure in duplicate.
8.1.2
Label two 1.5 mL microcentrifuge tubes for each sample, including QC strains (Step 7.1.)
8.1.3
Add 500 µL of 100% Ethanol to each.
8.1.4
Add approximately 50 µL of 0.1 mm diameter silica beads to each tube, using the scaling on the microcentrifuge tube.
8.1.5
Wet a sterile swab using the ethanol from the suspension (from step 8.1.3) or sterile deionized water.
8.1.6
Press swab against the side of the tube to remove excess fluid.
8.1.7
Collect approximately a circle of 1-2 cm diameter of mold from the agar plate using the swab, selecting spores (conidia) and hyphae if possible. Be careful not to pick any agar when picking up the colonies. NOTE: Each run, must include positive controls; Aspergillus fumigatus ATCC MYA-3626 and Aspergillus flavus ATCC MYA-3631 (or other validated molds) (See 7.1).
8.1.8
Suspend the collected material in the 1.5 mL tube.
8.1.9
Centrifuge for 3 minutes at 14,000 g.
8.1.10
Remove the Ethanol supernatant.
8.1.11
Re-suspend the pellet in 20 µL of 70% formic acid.
8.1.12
Vortex briefly.
8.1.13
Add 20 µL of acetonitrile.
8.1.14
Vortex briefly.
8.1.15
Centrifuge for 3 minutes at 14,000 g.
8.1.16
Add 1 μL of the supernatant, taking care not to disturb the pellet, and spot, in duplicate, to a 96-spot reusable stainless steel target plate (see Fig. 1)
8.1.17
Add 1 μL of the BTS suspension to two spots on the 96-spot reusable stainless steel target plate (do not add formic acid to BTS!) Simply allow to air dry and proceed to step 8.1.18.
Perform this step once a week. See step 4.7.1 for preparation of BTS.
8.1.18
As soon as spots are dry, immediately overlay with 1 μL of HCCA matrix. **Do not delay the addition of HCCA matrix as it will decrease the quality of your run.
8.1.19
Air dry for two minutes.
8.1.20
As soon as spots are dry, they are ready to be subject to MALDI-TOF analysis
NOTE: target must be completely dry before it is inserted into MALDI BioTyper.
8.1.21
Once the target plate is dry, the plate must be run within 24 hours.
8.1.22
Be sure that the location of each specimen on the 96-spot reusable stainless steel target plate is recorded on a sample key spreadsheet. Verify the spreadsheet numbers to make sure they match the original sample numbers.
8.1.23
Samples are now ready for the MALDI-ToF MS assay on the Bruker Biotyper platform
8.1.24
Resulting spectra must be interpreted using Bruker database.
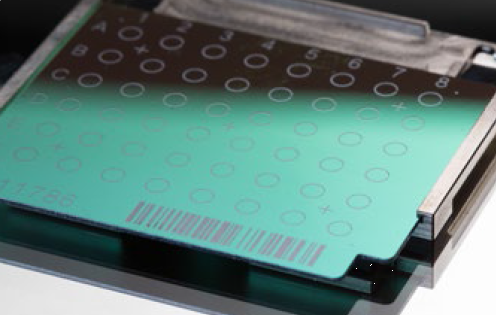
Figure 1. Example of stainless-steel target plate
10.1
Review Control Strains.
10.2
At least one of the duplicates for each control strain must generate a score of ≥ 1.7 with a correct genus level identification (no conflicting results between the duplicates).
10.2.1
If the control strains generate scores <1.7 at the genus level or an incorrect identification, the results cannot be interpreted, and the extraction/submission process must be repeated.
10.3
Mold species will be identified if database match scores are ≥2.0 for one or more of the duplicates and at the genus level only if scores fall between 1.7-1.9.
10.4
If no species identification is made, repeat extraction/submission process using a new subculture.
10.5
If no identification is successful after second submission, the isolate must be identified by another methodology.
11.1.1
Go to the MicrobeNet website microbenet.cdc.gov.
11.1.2
Request a MicrobeNet account.
11.1.3
Log into account.
11.1.4
Convert Main Spectra to XML file on your Bruker software.
11.1.5
Drag and drop the XML file onto the MicrobeNet website.
11.1.6
The top 10 matches will populate.
13.0 Disclaimer for electronically distributed standard operating procedures for CDC-developed tests:
The Mycotic Diseases Branch laboratory developed this document as an example test procedure for the identification of molds using MALDI-ToF. It is the responsibility of the testing laboratory to ensure content and format are modified as necessary to meet applicable regulatory requirements, quality management system standards, and chemical, radiological and biological safety requirements. This is not a controlled document and the described test methods are subject to change without notice. It is the responsibility of the testing laboratory to ensure the information within this document remains applicable. Contact the test developer at gyi2@cdc.gov to find out whether any changes have been made.
