Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus in Domestic Cat, France, 2022
François-Xavier Briand
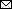
, Florent Souchaud, Isabelle Pierre, Véronique Beven, Edouard Hirchaud, Fabrice Hérault, René Planel, Angélina Rigaudeau, Sibylle Bernard-Stoecklin, Sylvie Van der Werf, Bruno Lina, Guillaume Gerbier, Nicolas Eterradossi, Audrey Schmitz, Eric Niqueux, and Béatrice Grasland
Author affiliations: ANSES, Ploufragan, France (F.-X. Briand, F. Souchaud, I. Pierre, V. Beven, E. Hirchaud, N. Eterradossi, A. Schmitz, E. Niqueux, B. Grasland); Clinique Vétérinaire des Deux Rivières, Mauléon, France (F. Hérault); Clinique Vétérinaire Filiavet, Bressuire, France (R. Planel); Resalab Ouest site de Labovet Analyse, Les Herbiers, France (A. Rigaudeau); Santé publique France, Saint-Maurice, France (S. Bernard-Stoecklin); Université Paris Cité Institut Pasteur National Reference Center, Paris, France (S. Van der Werf); National Reference Center for Respiratory Viruses, Lyon, France (B. Lina); Université de Lyon, Lyon (B. Lina); French Ministry of Food and Agriculture, Paris (G. Gerbier)
Main Article
Figure

Figure. Phylogenetic analysis of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus detected in domestic cat, France, 2022. Tree was created by using MEGA 7 software (https://megasoftware.net) and the neighbor-joining method with 1,000 bootstrap replicates for complete concatenated HPAI H5N1 virus segments. All sequences belong to the A/duck/Saratov/29-02V/2021–like genotype. Red solid circle indicates virus sequence from cat; black solid circle indicates sequence from a nearby duck farm. Both sequences are available in the GISAID database (https://www.gisaid.org) under accession nos. EPI_ISL_16395206 (cat) and EPI_ISL_16740903 (duck). Red bracket indicates closely related sequences detected during the same period and area in France from domestic bird farms. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: June 16, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
