Genomic Evidence of SARS-CoV-2 Reinfection Involving E484K Spike Mutation, Brazil
Carolina K. V. Nonaka, Marília Miranda Franco, Tiago Gräf, Camila Araújo de Lorenzo Barcia, Renata Naves de Ávila Mendonça, Karoline Almeida Felix de Sousa, Leila M. C. Neiva, Vagner Fosenca, Ana V. A. Mendes, Renato Santana de Aguiar, Marta Giovanetti
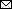
, and Bruno Solano de Freitas Souza
Author affiliations: D’Or Institute for Research and Education (IDOR), São Rafael Hospital Center for Biotechnology and Cell Therapy, Salvador, Brazil (C.K. Vasques Nonaka, B. Solano de Freitas Souza); São Rafael Hospital Department of Infectology, Salvador (M.M. Franco, C. Araújo de Lorenzo Barcia, R. Naves de Ávila Mendonça, K. Almeida Felix de Sousa, A. Verena Almeida Mendes); Fundação Oswaldo Cruz, Gonçalo Moniz Institute, Salvador (T. Gräf); Coordenação Geral dos Laboratórios de Saúde Pública/Ministério da Saúde, Brasília, Brazil (V. Fosenca); University of KwaZulu-Natal College of Health Sciences, Durban, South Africa (V. Fosenca); Universidade Federal de Minas Gerais Instituto de Ciências Biológicas, Belo Horizonte, Brazil (V. Fosenca, R.S. de Aguiar, M. Giovanetti); Fundação Oswaldo Cruz Instituto Oswaldo Cruz, Rio de Janeiro (M. Giovanetti); University Campus Bio-Medico of Rome Unit of Medical Statistics and Molecular Epidemiology, Rome, Italy (M. Giovanetti)
Main Article
Figure

Figure. Molecular characterization of a severe acute respiratory syndrome coronavirus 2 reinfection case in Salvador, Bahia State, northeast Brazil. A) Timeline of symptom onset and molecular and serologic diagnosis. B) Time-scaled maximum-likelihood tree, including the new genomes (GISAID accession nos. EPI_ISL_756293 and EPI_ISL_756294; https://www.gisaid.org) recovered from a 45-year-old woman residing in Salvador and full-length viral genomes from Brazil available through GISAID as of January 14, 2021 (Appendix Table, https://wwwnc.cdc.gov/EID/article/27/5/21-0191-App1.xlsx). New genomes are highlighted with red circles. Branch support (SH-aLTR >0.8) is shown at key nodes. C) Mutational pattern of the 2 isolates obtained from the same patient within a 147-day interval. Only unique mutations and lineage defining mutations for B.1.1.33 and P.2 are shown. ORF, open reading frame; rRT-PCR, real-time reverse transcription PCR; UTR, untranslated region.
Main Article
Page created: February 17, 2021
Page updated: April 22, 2021
Page reviewed: April 22, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
