Evolution and Antigenic Drift of Influenza A (H7N9) Viruses, China, 2017–2019
Jiahao Zhang
1, Hejia Ye
1, Huanan Li
1, Kaixiong Ma, Weihong Qiu, Yiqun Chen, Ziwen Qiu, Bo Li, Weixin Jia, Zhaoping Liang, Ming Liao
, and Wenbao Qi
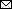
Author affiliations: College of Veterinary Medicine, South China Agricultural University, Guangzhou, China (J. Zhang, H. Li, K. Ma, Y. Chen, Z. Qiu, B. Li, W. Jia, M. Liao, W. Qi); National Avian Influenza Para-Reference Laboratory, Ministry of Agriculture and Rural Affairs of the People’s Republic of China, Guangzhou (J. Zhang, H. Li, W. Jia, M. Liao, W. Qi); Key Laboratory of Animal Vaccine Development, Ministry of Agriculture and Rural Affairs of the People’s Republic of China, Guangzhou (J. Zhang, W. Jia, M. Liao, W. Qi); National and Regional Joint Engineering Laboratory for Medicament of Zoonoses Prevention and Control, National Development and Reform Commission, Guangzhou (J. Zhang, W. Jia, M. Liao, W. Qi); Key Laboratory of Zoonoses Prevention and Control of Guangdong Province, Guangzhou (J. Zhang, W. Jia, M. Liao, W. Qi); Guangzhou South China Biological Medicine Co., Ltd, Guangzhou (H. Ye, W. Qiu, Z. Liang); Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou (W. Jia, M. Liao, W. Qi)
Main Article
Figure 1

Figure 1. Evolutionary history of influenza A(H7N9) viruses, China, 2017–2019. A) Phylogenic tree of the hemagglutinin gene of H7N9 viruses. Colors indicate reference H7N9 viruses (n = 1,038) from each wave together with the H7N9 isolates from this study (panel B). Red on the right of the tree indicates isolates from humans. All branch lengths are scaled according to the numbers of substitutions per site. The tree was rooted by using A/Shanghai/1/2013(H7N9), which was collected in February 2013. B) Hemagglutinin gene tree revealing a single cluster of highly pathogenic H7N9 viruses circulating during 2019. Red indicates the H7N9 isolates from this study. Scale bar represents number of nucleotide substitutions per site. C) Distribution of highly pathogenic influenza A(H7N9) viruses during 2019. The backgrounds indicate the sampling spaces of highly pathogenic influenza A(H7N9) viruses during 2019 in humans (red), environment (gray), and chickens (blue). The map was designed by using ArcGIS Desktop 10.4 software (ESRI, http://www.esri.com).
Main Article
Page created: June 12, 2020
Page updated: July 19, 2020
Page reviewed: July 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
