 |
|
|
|
|
|
|
| ||||||||||
|
|
|
|
|
|
|
||||
| ||||||||||
|
|
|
|
|
Persons using assistive technology might not be able to fully access information in this file. For assistance, please send e-mail to: mmwrq@cdc.gov. Type 508 Accommodation and the title of the report in the subject line of e-mail. Information System Architectures for Syndromic SurveillanceWilliam B. Lober, L. Trigg, B. Karras
Corresponding author: William B. Lober, University of Washington, MS 357240, 1959 NE Pacific St., Seattle, Washington 98195. Telephone: 206-616-6685; Fax: 206-338-2455; E-mail: lober@u.washington.edu. AbstractIntroduction: Public health agencies are developing the capacity to automatically acquire, integrate, and analyze clinical information for disease surveillance. The design of such surveillance systems might benefit from the incorporation of advanced architectures developed for biomedical data integration. Data integration is not unique to public health, and both information technology and academic research should influence development of these systems. Objectives: The goal of this paper is to describe the essential architectural components of a syndromic surveillance information system and discuss existing and potential architectural approaches to data integration. Methods: This paper examines the role of data elements, vocabulary standards, data extraction, transport and security, transformation and normalization, and analysis data sets in developing disease-surveillance systems. It then discusses automated surveillance systems in the context of biomedical and computer science research in data integration, both to characterize existing systems and to indicate potential avenues of investigation to build systems that support public health practice. Results: The Public Health Information Network (PHIN) identifies best practices for essential architectural components of a syndromic surveillance system. A schema for classifying biomedical data-integration software is useful for classifying present approaches to syndromic surveillance and for describing architectural variation. Conclusions: Public health informatics and computer science research in data-integration systems can supplement approaches recommended by PHIN and provide information for future public health surveillance systems. IntroductionAutomated acquisition of routine health-care data has enhanced public health surveillance capabilities. The 2003 National Syndromic Surveillance Conference featured model syndromic surveillance systems, including New York City's emergency department (ED)-based syndromic surveillance system (1), the Real-Time Outbreak Disease Surveillance system (RODS) (2), the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE) (3), and other encounter-based systems. These systems use different data sources, including ED and primary care outpatient data (e.g., chief complaints or diagnoses), diagnosis-specific aggregate data (National Bioterrorism Syndromic Surveillance Demonstration Project [4]), and laboratory and radiology data, for early detection of disease outbreaks. Surveillance to detect clinical syndromes, whether inferred by secondary use of clinical data sources or directly coded by observers, is commonly called syndromic surveillance. Despite increasingly widespread development of syndromic surveillance systems, continued efforts to understand different data-analysis strategies, and ongoing discussion of strategies to integrate syndromic surveillance into public health practice, the cost-benefit ratio of syndromic surveillance remains uncertain. Recommendations from the 2001 American Medical Informatics Association meeting stated that "public health informatics must create an information architecture that includes a longitudinal, person-based, integrated data repository…similar to the National Electronic Disease Surveillance System (NEDSS) model" (5). NEDSS has evolved into a prominent component of the Public Health Informatics Network (PHIN) initiative (6,7). A recent review of PHIN (8) concluded that "the PHIN vision must continue to broaden beyond the structured data obtained from surveillance systems and labs to include syndromic data from clinics, ERs, doctor's offices, pharmacies…," indicating that surveillance based on integration of heterogeneous data will become central to public health practice. Implementing syndromic surveillance based on automated acquisition of clinical data requires both the development of secure, reliable information systems and the use of those systems in public health practice. The information technology (IT) activities include system design and integration and development of tools for data acquisition and analysis. Effective use of syndromic surveillance depends not only on IT activities but also on the system's integration with public health practices for outbreak detection, investigation, and response management. Data modeling and data integration are integral IT components of syndromic surveillance information systems. Data modeling activities are those related to structure and content, and entail identifying relevant clinical variables; understanding both the vocabularies and coding schemes used to record these variables; and establishing procedures for clustering, re-coding, normalizing, or otherwise preparing data for analysis. Data integration activities are those related to movement and processing of data before their analysis or visualization, and entail acquiring, transforming, storing, and delivering information securely and reliably. Approaches to data modeling and integration and the trade-offs between different implementation technologies constrain the choice of system architectures. This paper reviews these components in the context of basic and applied research in data integration, on the basis of an evolutionary model used to describe the development of biomedical informatics (9). This model provides a framework for reviewing architectures used for automated public health surveillance, both to classify them and to discuss the strengths, weaknesses, and roles of different research approaches. Data Model ComponentsLimited development of syndromic surveillance systems, including the RODS and Syndromic Surveillance Information Collection (SSIC) systems (10), occurred before the anthrax outbreak in fall 2001. However, the 2001 terrorist attacks precipitated an increase in syndromic surveillance development, and implementation since then has balanced standardization with expediency. To implement systems rapidly before another terrorist attack, developers built systems tailored to readily available data. However, promulgation of national standards (e.g., PHIN) has emphasized the need for standardization of data types collected and of vocabularies used for individual data elements. Data ElementsTwo important data-element considerations are 1) the composition of the extracted data set and 2) the level of identification of the data. A 2001 review of data elements collected for surveillance by 10 different systems identified striking similarities (11). The majority of systems described at the 2003 NSSC continue to use data elements identified by that review. These systems collect data for patient ED or primary care visits and typically include age, sex, visit date and time, a measure of chief complaint and/or diagnosis, and a geographic measure; however, data elements and coding schemes vary among systems. Chief complaints or diagnoses, clustered into syndrome groupings, are used as variables for analysis, and both demographic and geographic variables are used to stratify the data. In contrast to the simple data model used by the majority of syndromic surveillance systems, the PHIN Logical Data Model provides a rich, detailed, object-oriented view of health-care data (12), encouraging both standardization and more granular data collection. Public health agencies have legal authority to collect (the minimum necessary) data for surveillance, "without [patient] authorization, for the purpose of preventing or controlling disease, injury, or disability…" (13). However, certain barriers to provider data reporting have been identified, including regulatory issues, fit with business model, use of IT resources, public relations, accounting for public health disclosures, and release of competitive data (14). Despite certain states' legal authority to collect identified data, multiple system developers have chosen to collect either de-identified or minimally identified data to reduce these practical barriers. Although a masked or encrypted identifier can address these concerns while maintaining data quality, this approach was challenged by the final interpretation of the Health Information Portability and Accountability Act of 1996 (HIPAA) Privacy Rule (15). Concern has been expressed about the effect of this interpretation on medical and public health research (16). Standard Vocabulary UsageStandards for exchange of public health data arose from the need to combine heterogeneous data (i.e., comparable clinical information from different sources that is expressed by using different formats and coding schemes). PHIN specifies the ability to translate and manipulate Logical Observation Identifiers Names and Codes (LOINC®), Systematized Nomenclature of Medicine (SNOMED®), International Classification of Diseases, Ninth Revision (ICD-9), and current procedural terminology (CPT®) codes and to map local, legacy, or proprietary codes into these standards (Table). PHIN specifies LOINC as the vocabulary for laboratory reporting in conjunction with the PHIN notifiable-condition--mapping tables (17), which map LOINC and SNOMED codes to reportable conditions. Unlike laboratory reporting, syndromic surveillance systems might use local vocabularies and lack a fully developed transformation capability. Hospitals use different standard coding schemes, and transformation will become increasingly important as the scale of these systems increases. Analysis Data ModelAggregating data for analysis is also a challenge. Systems commonly use ICD-9 codes or chief-complaint data to categorize illnesses into syndrome groups. Different ICD-9 clustering schemes exist, including a collaborative effort of CDC and other agencies (18). Assigning chief complaints to syndrome groups has been implemented in different ways, including by Bayesian classification (19) and text substring searches (2), and is still being studied. Current algorithms and statistical approaches to detection have been implemented either by using standard statistical software packages or as part of the surveillance system. In either case, the information-system architecture should support preparation of an analysis data set by using a model appropriate to its intended use, the secure delivery of the data to the algorithm, and the data analysis and results presentation itself. Data-Integration ComponentsData integration is characterized by five functions: data extraction, secure data transport, transformation, normalization, and creation of an analysis data set or view. Systems use different approaches to perform these functions; PHIN cites multiple best practices. Data ExtractionData extraction refers to acquiring a data set from the source system. Query-based systems extract data through periodic execution of local queries or reports. IT staff responsible for the source system often develop these queries and run them automatically. In certain circumstances, queries against the source system are executed directly by the surveillance system. Message- or event-based systems send a message to the surveillance system whenever something of interest occurs in the source system. Typically, this stream of messages contains either the entire message set, or a filtered subset, of an electronic data interchange between hospital systems. These messages are commonly in Health Level 7 (HL7) format (20) and often can be rerouted by using the hospitals' HL7 interface engine or message switchboard. PHIN refers to a series of standards, including HL7 2.x and 3.0, to describe the appropriate formatting for data sent from a source system to public health authorities. However, both query-based and message-based data are consistent with PHIN. Transport and SecurityPublic health surveillance data typically travel through the Internet. Although the chance of data either being intercepted or spoofed is low, certain techniques can ensure encryption of the message and protection of participants' identities (21). Files can be encrypted and signed by using a standard (e.g., Pretty Good Privacy [PGP]), transferred through a virtual private network (VPN), or transmitted by using a file transfer protocol (FTP) over a securely encrypted channel. PHIN specifies the PHIN Messaging System (PHINMS), which is based on ebXML standard for bidirectional data transport. Symmetric public key encryption (PKI), in which both parties use X.509 certificates, offers both high-quality channel encryption and authentication of both sides of the conversation and is used by PHINMS. PHIN also recommends annual security evaluation. Transformation and NormalizationData arriving from different source systems can be in different formats, and coding schemes used for individual data elements might need to be reconciled. Transformation of syntax and normalization of semantics must be organized and well-documented. The complexity of these steps is a direct result of the variance among the source systems. A trade-off exists between the complexity of programming needed to manage these transformations and the complexity of the human relationships needed to ensure that formats are synchronized among separate institutions. Certain systems represent data by using extensible markup language (XML), thus allowing data to be manipulated through standard transformation parsers. PHIN specifies use of XML and the need for a data-translation capability, without specifying software packages or platforms. Analysis Set Creation and DeliveryFinally, integrated and normalized data need to be presented for analysis. The performance of detection algorithms is being researched, and the needs of different detection algorithms vary (22). Even when using a specific algorithm, users might not know whether to count each patient as a single data point or allow multiple data points for a patient who meets criteria for multiple syndromes. A flexible query system can present multiple analysis sets either to a human or to an automated detection algorithm. PHIN calls for the capability to analyze, display, report, and map data. These features are implemented in the model systems, but not always with comparable algorithms. Architectures for Data IntegrationThe challenges of integrating data from heterogeneous sources into a single analysis set are not unique to public health surveillance. Decision support in business endeavors often depends on integrating and analyzing diverse data sets. Clinical practice increasingly requires this capability, as patient information is often widely distributed and patient care requires access to information at other institutions. This need to access distributed information is central to automated public health surveillance. Three generations of data-integration techniques in biomedical informatics have been described (10). The simplest approach to data integration is to build a large-source system containing all data needed to satisfy a query. As data have multiplied, along with their diversity, uses, and ownership considerations, new integration approaches have been developed. Second-generation models integrate data from multiple sources at a central location. This technology is almost universal in public health surveillance systems. A third-generation approach is emerging that involves constructing relations between data sources so that they appear integrated to the surveillance user, even though the data remain at their original location, subject to the control of their original owner. Distinct models for this third-generation approach exist; research in this area has only recently been applied to public health surveillance. First-Generation IntegrationSurveillance based on first-generation systems is not practical unless a single information system contains sufficient data to represent the population of interest. A slight enhancement is the manual combination of data from multiple nonintegrated sources. This is often a first step in local public health surveillance. A health department might receive files containing surveillance reports and combine them manually by using a spreadsheet program, desktop database, or statistical data management package. This approach is straightforward but can result in data fields with cryptic, local meanings and in data elements represented in a combination of nonuniform coding schemes from different sources. Second-Generation IntegrationSecond-generation integration has been characterized as the consolidation of data through enterprise information architecture (10). One sophisticated second-generation approach is data warehousing, characterized as "historical, summarized, and consolidated data… targeted for decision support" (23). Data warehousing systems are common in business and widely available in health care. Their characteristics closely match those desired for public health surveillance, although data are less timely than desired. Warehouse data are typically historic; although historic data can be useful for research and for developing event-detection algorithms, ongoing surveillance requires current data. At present, the common model for automated public health surveillance systems is a data warehouse with frequent updates (although the term warehouse is not typically used in syndromic surveillance literature). Although multiple approaches to data warehousing exist, all approaches are implemented through construction of a centralized database that is optimized for resource-intensive queries against a substantial portion of data. Data from other sources are typically imported to the central warehouse database after a query is sent from the central database to the source database. A lag associated with periodic imports from the clinical database(s) into the warehouse is commonplace and has been noted in multiple query-based public health surveillance systems (2,4,5,11). Another limitation of this model is that a global schema, or data structure, is required, and a change in this schema typically requires changes in the import procedures from each source system. One variant of this approach is for data sources to run local queries and transmit resulting data on a schedule. A second variant involves filtering the electronic data interchange messages used to transfer data between components of an enterprise clinical information system and storing data contained therein. These data are usually formatted according to the HL7 standard. This approach can improve data timeliness substantially, and the uniformity of HL7 encoding might simplify software development. However, substantial variation is permitted within the standard, and HL7-message decoding often requires customization. Moreover, this approach requires a consistent global schema and the mapping of that schema into multiple local variations. This approach is exemplified by RODS (2). Third-Generation IntegrationThe need for future information systems to support automated information acquisition processes for decision-making activities was identified over a decade ago (24). To achieve this automation, an approach based on so-called mediators (i.e., software agents that translate a query from a global format to an appropriate local format for a specific database) was proposed. This model, in which queries are run against distributed, in-situ data, can be classified as a third-generation data-integration system. Contemporary approaches to third-generation systems include federated databases, mediated query systems, and peer-to-peer data sharing. These approaches share an apparent integration of information that remains housed in multiple source systems. Although these systems might be more complex to build than earlier generation systems, they share the advantage that data are queried from their original location, which improves accuracy and decreases lag. Additionally, lightweight queries can be run routinely for surveillance purposes with less performance impact on the source system. More richly detailed underlying data might be equally available for focused investigations. However, early third-generation approaches expose the source system to performance degradation from additional queries and require that the source system be online to process a query. Ongoing research is aimed at minimizing these shortcomings and strengthening the approaches' advantages. Federated databases are an association of independent databases that allow queries from a single source but have no common schema or organization (25). The lack of a common schema means that any application must contain the local schema for every database it wishes to query. This is efficient because the queries generated require no translation for the source systems, but each new data source added to the system might require a change in each application accessing data from the federation. A number of federated-database models exist; these differ in the locus and degree of centralized control over access to the system (26). The Kleisli system is one federated-database approach for integrating bioinformatics data, in which a set of drivers provides access to various heterogeneous databases (27). One proposed mediator query model has been implemented in biomedical applications, which again provide integrated access to online genetic databases (24). Examples include the Biomediator (28) and transparent access to multiple bioinformatics information sources (TAMBIS) (29) systems. Mediated schema models offer real-time queries directly against source systems, combined with the single global schema of a data warehouse. This greatly simplifies application writing, as authors need to understand only the single common schema. This model has not yet been implemented in public health surveillance. Perhaps the best-known applications of peer-to-peer communication are music- and file-sharing services (e.g., Napster and Gnutella). These services use somewhat different peer-to-peer models, using a common schema but maintaining their common index information in either a centralized or distributed fashion, respectively. This peer-to-peer file-sharing model, extended to include peered communication among intelligent data-sharing agents, has been described as a peer-data--management system (30). Although third-generation systems are not widespread in public health surveillance, these models are promising. First, whether executed through a mediated schema or against a series of autonomous peer agents, the queries in these systems run directly against the source data. Timeliness and accuracy are ensured, and performance concerns can be mitigated by different strategies. These architectures are suitable to run against both modern and legacy databases, transparently presenting an integrated view of both. The intelligence built into each participant of a peer-data--management system lends itself to supporting queries that can dynamically configure themselves against the available data sources when they are run. Finally, local control over data sources, which is inherent in both peer-data--management systems and the mediated-schema approach, might enable owners of data at any level to provide access and detail appropriate to different stakeholders and in different situations while maintaining control of their own data. ConclusionsIn response to the threat of biologic terrorism, information-system--based public health surveillance has evolved rapidly. Second- and third-generation approaches offer the greatest utility for public health surveillance, and research is critical to the continued advancement of surveillance systems, especially third-generation systems. Future research on surveillance architectures should explore combinations of methods. For example, a data warehouse that provides the data consolidation, rich historic record, comprehensible data structure, and ability to query the entire corpus of data at the local public health jurisdiction might be combined with a third-generation model for sharing of situation-dependent views of those data. The nature of this integration will be driven by issues of data ownership and privacy, as well as by an evolving understanding of the optimal data for various uses. Broad-scale application of these systems will also require policy development to address concerns of privacy and proprietary data. Public health agencies should partner with universities and research organizations to shape the agenda for data-integration research. At the same time, academics need public health partners to ensure that research questions are grounded in relevant problems. In addition, public health agencies must rely on proven technologies for their operational needs. This implies working with information system vendors to take advantage of data-integration solutions and to ensure those solutions meet public health needs. As has been the case in clinical informatics, where data used for outcomes research are also useful for chronic disease management, quality assurance, health services research, and other purposes, surveillance systems likely will evolve to enhance public and environmental health practice and management. Public health leaders should pay attention to how these data-integration models scale in other domains; links with the research community will prove helpful. Although public health agencies must serve an immediate operational role in national security, aggressive research is required to extend the frontiers of data integration to ensure success. Acknowledgments Peter Mork, University of Washington Department of Computer Science and Engineering, provided a thorough review of the architecture section. References
Table 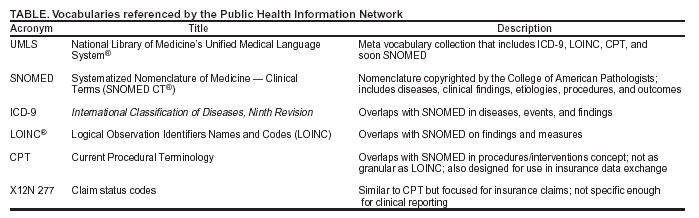 Return to top.
Disclaimer All MMWR HTML versions of articles are electronic conversions from ASCII text into HTML. This conversion may have resulted in character translation or format errors in the HTML version. Users should not rely on this HTML document, but are referred to the electronic PDF version and/or the original MMWR paper copy for the official text, figures, and tables. An original paper copy of this issue can be obtained from the Superintendent of Documents, U.S. Government Printing Office (GPO), Washington, DC 20402-9371; telephone: (202) 512-1800. Contact GPO for current prices. **Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.Page converted: 9/14/2004 |
|||||||||
This page last reviewed 9/14/2004
|