Model Performance Evaluation Program Report of Results: August 2021
MPEP August 2021 pdf icon[PDF – 1 MB]
The purpose of this report is to present results of the U.S. Centers for Disease Control and Prevention (CDC) Model Performance Evaluation Program (MPEP) for Mycobacterium tuberculosis complex (MTBC) drug susceptibility testing survey sent to participants in August 2021.
Primary Classification
This report contains DST results submitted to CDC by survey participants at 68 laboratories in 34 states.
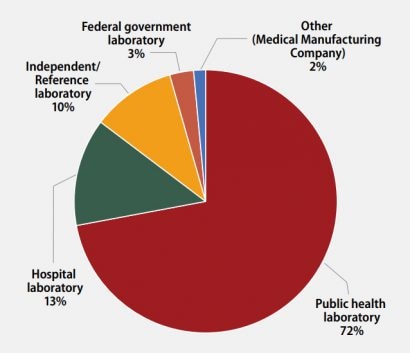
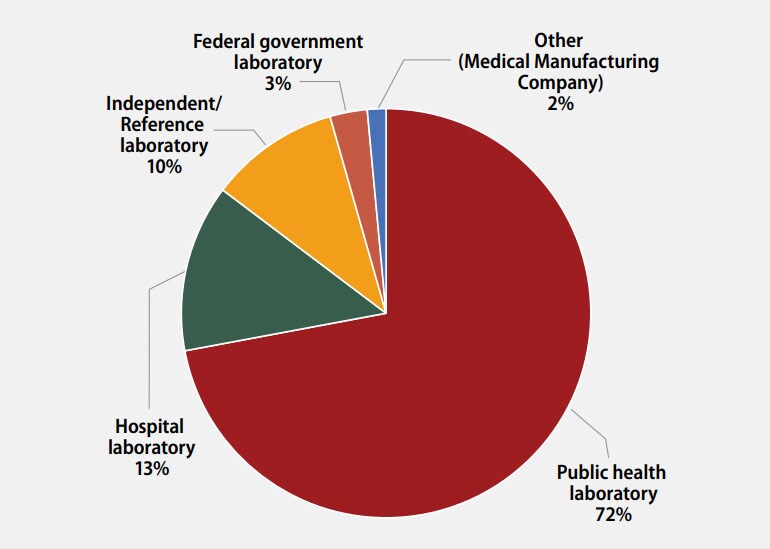
The participants were asked to indicate the primary classification of their laboratory (Figure 1). MPEP participants self-classified as:
- 49 (72%): Public health laboratory (e.g., local, county, state)
- 9 (13%): Hospital laboratory
- 7 (10%): Independent/Reference laboratory (non-hospital based)
- 2 (3%): Federal government laboratory
- 1 (2%): Other (Medical Manufacturing Company)
Annual Number of MTBC Drug Susceptibility Tests Performed
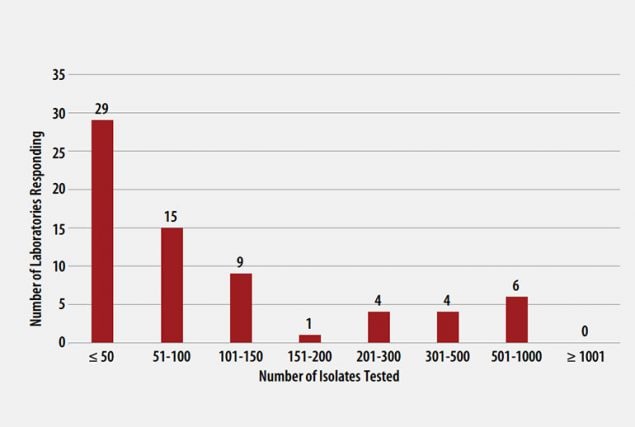
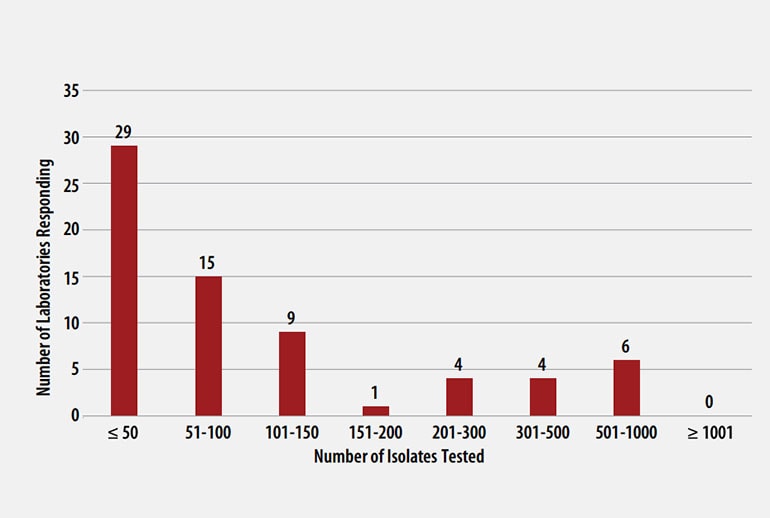
The number of MTBC isolates tested for drug susceptibility by the 68 participants in 2020 (excluding isolates used for quality control) is shown in Figure 2. In 2020, the counts ranged from 0 to 782 tests. Participants at 29 (42%) laboratories reported testing 50 or fewer DST isolates per year. Laboratories with low MTBC DST volumes are encouraged to consider referral of testing because of concerns about maintaining proficiency [8].
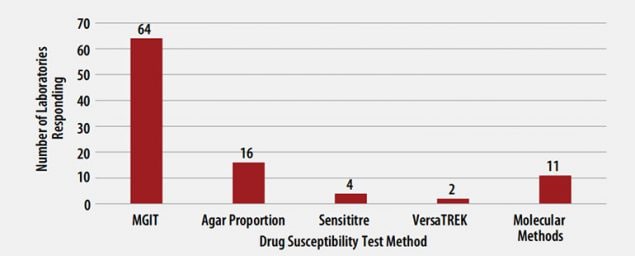
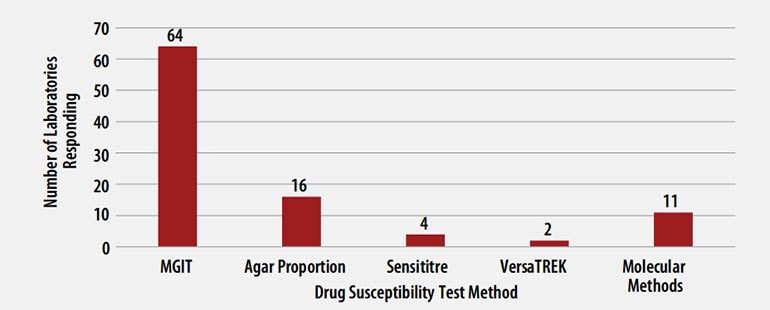
MTBC DST Methods Used by Participants
The DST methods that were used by participating laboratories for this panel of MTBC isolates are displayed in Figure 3. Of participating laboratories, 43 (63%) reported results for only one method, 21 (31%) reported two methods, and 4 (6%) noted three susceptibility methods.
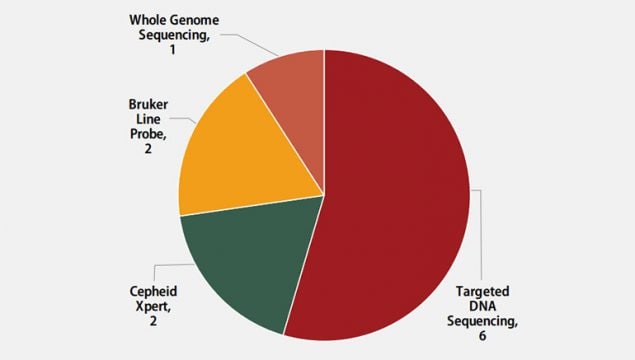
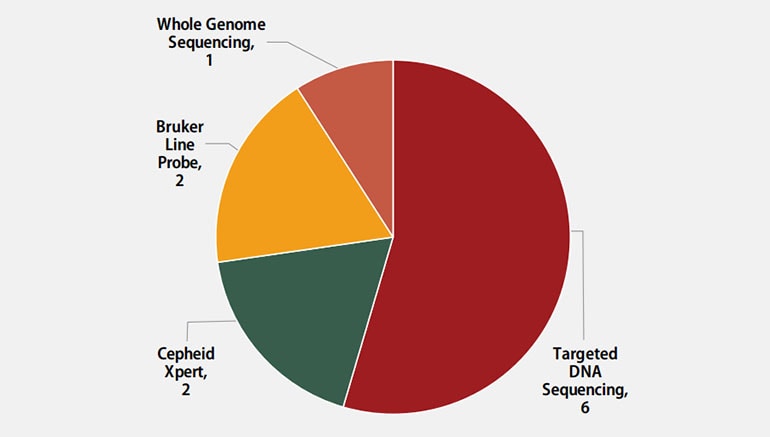
Molecular methods reported by participants are shown in Figure 4. The method used most frequently by six laboratories (55%) was targeted DNA sequencing, including pyrosequencing and Sanger sequencing. Two (18%) laboratories reported use of the Cepheid Xpert MTB/RIF assay, two (18%) reported results for line probe assays, Bruker Genotype MTBDRplus and MTBDRsl, and one (9%) reported results from whole genome sequencing.
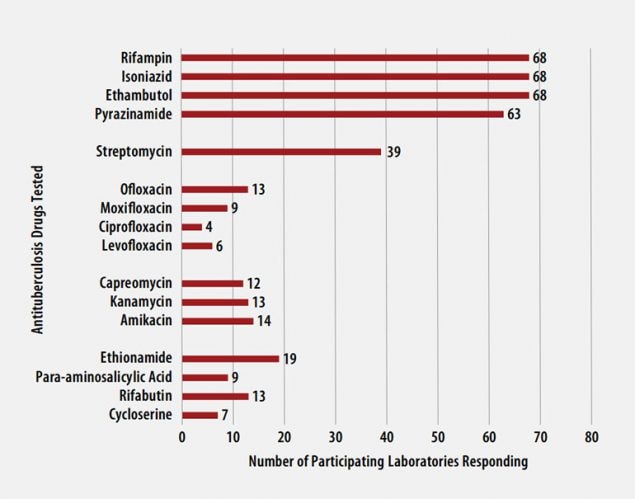
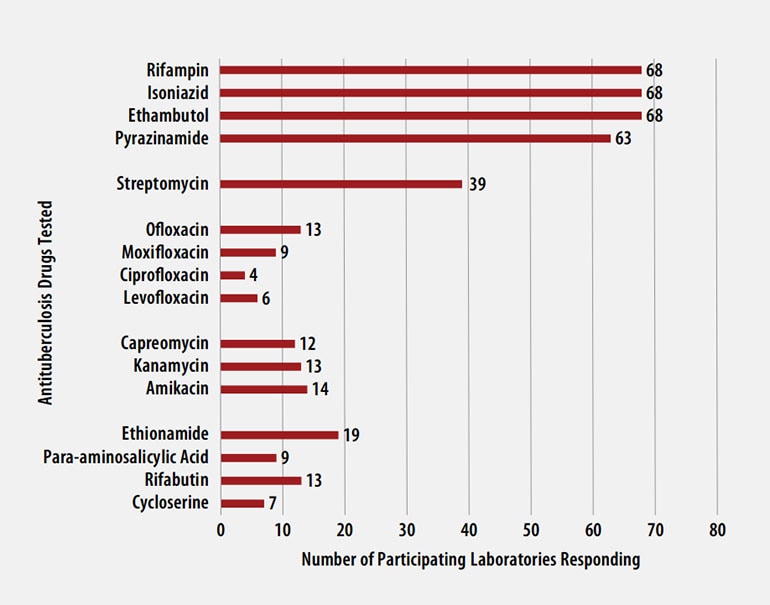
Antituberculosis Drugs Tested by Participants
The number of participating laboratories that reported testing each antituberculosis drug in the August 2021 survey is presented in Figure 5. CLSI recommends testing a full panel of first-line drugs (rifampin [RMP], isoniazid [INH], ethambutol [EMB] and pyrazinamide [PZA])[1] because it represents a combination of tests that provides the clinician with comprehensive information related to the four-drug antituberculosis therapy currently recommended for most patients. All participants reported results for three of the first-line drugs (RMP, INH, and EMB) and 63 (93%) also reported results for PZA by growth-based DST methods. One laboratory performs molecular testing for PZA via sequencing of pncA, in place of growth-based DST.
For 21 laboratories reporting second-line drug results (with the exception of streptomycin), five (22%) tested all three second-line injectable drugs (amikacin, kanamycin, and capreomycin) and at least one fluoroquinolone (ofloxacin, ciprofloxacin, levofloxacin, or moxifloxacin) needed to confidently define XDR TB. CDC has adopted a new hybrid definition of XDR that includes both the former classification (i.e., MDR with resistance to second-line injectable plus fluoroquinolone) or the revised WHO definition (i.e., MDR plus resistance to fluoroquinolone and either bedaquiline or linezolid) [9, 10].
Anticipated growth-based and molecular results for the panel of MTBC isolates sent to participants in August 2021 are shown in the tables below. Although CDC recommends broth-based methods for routine first-line DST of MTBC isolates, the results obtained by the reference agar proportion method (except for pyrazinamide, in which MGIT was performed) are shown in Table 1. Molecular results obtained by DNA sequencing are listed in Table 2 [5].
Table 1. Expected Growth-based Results for August 2021 Survey
Note—S=susceptible, R=resistant
| Isolate | RMP | INH | EMB | PZA | Second-line Drugs Resistances: |
| 2021F | S | S | S | S | OFL, CIP |
| 2021G | S | R | S | S | STR, OFL, CIP |
| 2021H | S | R | S | S | OFL, CIP, ETA |
| 2021I | S | S | S | S* | OFL, CIP |
| 2021J | S | R | S | S | OFL, CIP, ETA |
*80% consensus for a single categorical result for this drug of either susceptible or resistant was not achieved for this isolate among participating laboratories.
Table 2. Expected Molecular Results (Mutations Detected in Loci Associated with Resistance) for August 2021 Survey
Note—Empty cell=No mutation detected
| Isolate | rpoB¥ | katG | inhA | fabG1 | gyrA |
|---|---|---|---|---|---|
| 2021F | Ala90Val | ||||
| 2021G | Asp94Asn | Asp94Gly | |||
| 2021H | Arg447Arg* (Arg528Arg)† |
Leu203Leu | Asp94Asn | ||
| 2021I | Ser91Pro | ||||
| 2021J | C-15T | Ala90Val |
¥Mutation is listed using both the M. tuberculosis and E.coli numbering system [6, 7]
*M. tuberculosis numbering system used
†E. coli numbering system used
- CLSI, Susceptibility Testing of Mycobacteria, Nocardiae spp., and Other Aerobic Actinomycetes, in 3rd Ed. CLSI Standard M24. 2018, Clinical and Laboratory Standards Institute: Wayne, PA.
- CLSI, Performance Standards for Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes, in 1st Ed. CLSI supplement M62. 2018, Clinical and Laboratory Standards Institute: Wayne, PA.
- World Health Organization, Technical Report on critical concentrations for drug susceptibility testing of isoniazid and the rifamycins (rifampicin, rifabutin and rifapentine). 2021: Geneva.
- World Health Organization, Technical Report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. 2018: Geneva.
- Campbell, P.J., et al., Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother, 2011. 55(5): p. 2032-41.
- Andre, E., et al., Consensus numbering system for the rifampicin resistance-associated rpoB gene mutations in pathogenic mycobacteria. Clin Microbiol Infect, 2017. 23(3): p. 167-172.
- APHL, Issues in Mycobacterium tuberculosis complex (MTBC) Drug Susceptibility Testing: Rifampin (RIF), in APHL Issues in Brief: Infectious Diseases. 2019, Association of Public Health Laboratories: Washington, D.C.
- APHL, TB Drug Susceptibility Testing Expert Panel Meeting Summary Report. 2007, Association of Public Health Laboratories: Washington, D.C.
- World Health Organization, Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis, 27-29 October 2020. 2021, World Health Organization: Geneva.
- CDC Division of Tuberculosis Elimination, Dear Colleague Letter: Surveillance definitions for extensively drug resistant (XDR) and pre-XDR tuberculosis. 2022.
- Devasia, R.A., et al., Fluoroquinolone resistance in Mycobacterium tuberculosis: the effect of duration and timing of fluoroquinolone exposure. Am J Respir Crit Care Med, 2009. 180(4): p. 365-70.
- Carr W, K.E., Starks A, Goswami N, Allen L, Winston C, Interim Guidance: 4-Month Rifapentine-Moxifloxacin Regimen for the Treatment of Drug-Susceptible Pulmonary Tuberculosis—United States, 2022. MMWR Morb Mortal Wkly Rep, 2022. 71(8): p. 285-289.
- Chen, T.C., et al., Fluoroquinolones are associated with delayed treatment and resistance in tuberculosis: a systematic review and meta-analysis. Int J Infect Dis, 2011. 15(3): p. e211-6.
- Zhang, Y. and W.W. Yew, Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis, 2015. 19(11): p. 1276-89.
- Zhang, Y. and W.W. Yew, Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis, 2009. 13(11): p. 1320-30.
- Eilertson, B., et al., High proportion of heteroresistance in gyrA and gyrB in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother, 2014. 58(6): p. 3270-5.
- Rinder, H., K.T. Mieskes, and T. Loscher, Heteroresistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis, 2001. 5(4): p. 339-45.
- Willby, M., et al., Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother, 2015. 59(9): p. 5427-34.
- Kam, K.M., et al., Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: correlation with ofloxacin susceptibility. Microb Drug Resist, 2006. 12(1): p. 7-11.
- Maruri, F., et al., A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. Journal of Antimicrobial Chemotherapy, 2012. 67(4): p. 819-831.
- Almeida Da Silva, P.E. and J.C. Palomino, Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother, 2011. 66(7): p. 1417-30.
- Seifert, M., et al., Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One, 2015. 10(3): p. e0119628.
- Kandler, J.L., et al., Validation of Novel Mycobacterium tuberculosis Isoniazid Resistance Mutations Not Detectable by Common Molecular Tests. Antimicrob Agents Chemother, 2018. 62(10).
- Ramaswamy, S.V., et al., Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother, 2003. 47(4): p. 1241-50.
- Ando, H., et al., A silent mutation in mabA confers isoniazid resistance on Mycobacterium tuberculosis. Mol Microbiol, 2014. 91(3): p. 538-47.
- Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. 2021, World Health Organization: Geneva.
- Morlock, G.P., et al., ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother, 2003. 47(12): p. 3799-805.
- Varma-Basil, M. and R. Prasad, Dilemmas with ethionamide susceptibility testing of Mycobacterium tuberculosis: A microbiologist & physician’s nightmare. Indian J Med Res, 2015. 142(5): p. 512-4.
- Centers for Disease Control and Prevention, Availability of an assay for detecting Mycobacterium tuberculosis, including rifampin-resistant strains, and considerations for its use – United States, 2013. MMWR Morb Mortal Wkly Rep, 2013. 62(41): p. 821-7.
- Van Deun, A., et al., Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol, 2013. 51(8): p. 2633-40.
- Ramirez-Busby, S.M. and F. Valafar, Systematic Review of Mutations in Pyrazinamidase Associated with Pyrazinamide Resistance in Mycobacterium tuberculosis Clinical Isolates. Antimicrob Agents Chemother, 2015. 59(9): p. 5267-77.
- Chedore, P., et al., Potential for erroneous results indicating resistance when using the Bactec MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol, 2010. 48(1): p. 300-1.