What is Rabies?
Rabies is a preventable viral disease most often transmitted through the bite of a rabid animal. The rabies virus infects the central nervous system of mammals, ultimately causing disease in the brain and death. The vast majority of rabies cases reported to the Centers for Disease Control and Prevention (CDC) each year occur in wild animals like bats, raccoons, skunks, and foxes, although any mammal can get rabies.
The Rabies Virus
Rabies virus belongs to the order Mononegavirales, viruses with a nonsegmented, negative-stranded RNA genomes. Within this group, viruses with a distinct “bullet” shape are classified in the Rhabdoviridae family, which includes at least three genera of animal viruses, Lyssavirus, Ephemerovirus, and Vesiculovirus. The genus Lyssavirus includes rabies virus, Lagos bat, Mokola virus, Duvenhage virus, European bat virus 1 & 2 and Australian bat virus.
Structure
Rhabdoviruses are approximately 180 nm long and 75 nm wide. The rabies genome encodes five proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and polymerase (L). All rhabdoviruses have two major structural components: a helical ribonucleoprotein core (RNP) and a surrounding envelope. In the RNP, genomic RNA is tightly encased by the nucleoprotein. Two other viral proteins, the phospoprotein and the large protein (L-protein or polymerase) are associated with the RNP.
The glycoprotein forms approximately 400 trimeric spikes which are tightly arranged on the surface of the virus. The M protein is associated both with the envelope and the RNP and may be the central protein of rhabdovirus assembly. The basic structure and composition of rabies virus is depicted in the longitudinal diagram below.
Rabies is an RNA virus. The genome encodes 5 proteins designated as N, P, M, G, and L. The order and relative size of the genes in the genome are shown in the figure below. The arrangement of these proteins and the RNA genome determine the structure of the rabies virus.
Replication
The fusion of the rabies virus envelope to the host cell membrane (adsorption) initiates the infection process. The interaction of the G protein and specific cell surface receptors may be involved.
After adsorption, the virus penetrates the host cell and enters the cytoplasm. The virions aggregate in the large endosomes (cytoplasmic vesicles). The viral membranes fuse to the endosomal membranes, causing the release of viral RNP into the cytoplasm (uncoating). Because lyssaviruses have a linear single-negative-stranded ribonucleic acid (RNA) genome, messenger RNAs (mRNAs) must be transcribed to permit virus replication.
A viral-encoded polymerase (L gene) transcribes the genomic strand of rabies RNA into leader RNA and five capped and polyadenylated mRNAs, which are translated into proteins. Translation, which involves the synthesis of the N, P, M, G and L proteins, occurs on free ribosomes in the cytoplasm. Although G protein synthesis is initiated on free ribosomes, completion of synthesis and glycosylation (processing of the glycoprotein), occurs in the endoplamsic reticulum (ER) and Golgi apparatus. The intracellular ratio of leader RNA to N protein regulates the switch from transcription to replication. When this switch is activated, replication of the viral genome begins. The first step in viral replication is synthesis of full-length copies (postive strands) of the viral genome. When the switch to replication occurs, RNA transcription becomes “non-stop” and stop codons are ignored. The viral polymerase enters a single site on the 3’ end of the genome, and proceeds to synthesize full-length copies of the genome. These positive strands of rabies RNA serve as templates for synthesis of full-length negative strands of the viral genome.
During the assembly process, the N-P-L complex encapsulates negative-stranded genomic RNA to form the RNP core, and the M protein forms a capsule, or matrix, around the RNP. The RNP-M complex migrates to an area of the plasma membrane containing glycoprotein inserts, and the M-protein initiates coiling. The M-RNP complex binds with the glycoprotein, and the completed virus buds from the plasma membrane. Within the central nervous system (CNS), there is preferential viral budding from plasma membranes. Conversely, virus in the salivary glands buds primarily from the cell membrane into the acinar lumen. Viral budding into the salivary gland and virus-induced aggressive biting-behavior in the host animal maximize chances of viral infection of a new host.
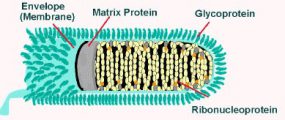
Rabies virions are bullet-shaped with 10-nm spike-like glycoprotein peplomers covering the surface. The ribonucleoprotein is composed of RNA encased in nucleoprotein -(), phosphorylated or phosphoprotein -Illistration of virus, and polymerase -virus.
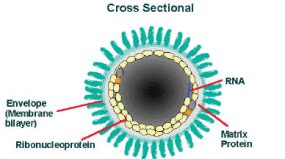
The cross-sectional diagram demonstrates the concentric layers: envelope membrane bilayer, M protein, and tightly coiled encased genomic RNA.

The rabies virus genome is single-stranded, antisense, nonsegmented, RNA of approximately 12 kb. There is a leader-sequence (LDR) of approximately 50 nucleotides, followed by N, P, M, G, and L genes.
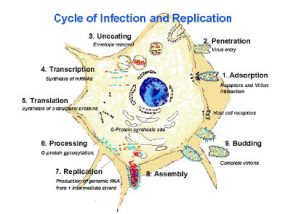
1: Adsorption (receptors and virion interation). 2: Penetration (virus entry). 3: Uncoating (envelope removal). 4. Transcription (synthesis of mRNAs). 5. Translation (Synthesis of structural proteins). 6. Processing (G-protein gycosylation). 7. Replication (production of genomic RNA from intermediate strand. 8. Assembly. 9: Budding (complete virions).

