AMD: Strengthening Food Safety
Each year, 1 in 6 Americans gets sick with a foodborne disease. To stop outbreaks of foodborne illness, investigators need to identify which pathogen is making people sick and then find the food source that is contaminated with the same pathogen. AMD methods, such as whole genome and next-generation sequencing (WGS and NGS) technologies, are helping link pathogens causing illnesses in people to contaminated food sources. Using AMD, CDC can more precisely identify the pathogens responsible for foodborne disease outbreaks.
Since 1996, PulseNet, a national laboratory network, has used bacterial DNA “fingerprinting” methods to connect foodborne illnesses to detect outbreaks. AMD technology is improving PulseNet’s ability to link illnesses to food sources. In 2013, CDC began helping PulseNet laboratories throughout the nation use AMD methods on foodborne bacteria, increasing the speed at which labs can detect outbreaks and identify contaminated food sources. By removing contaminated food sources from store shelves and out of people’s homes faster, public health programs can prevent additional illnesses.
As of March 2019, 69 laboratories in 49 states were PulseNet lab certified for whole genome sequencing of four major foodborne bacteria—Salmonella, Listeria, Shiga toxin-producing E. coli and Campylobacter.
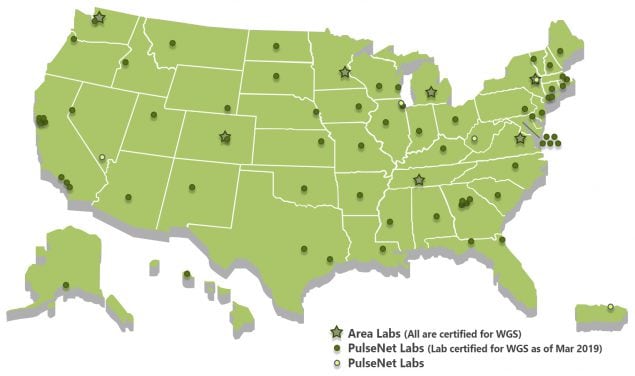
Read more about PulseNet participants here.
Norovirus is the leading cause of illnesses from contaminated food in the United States, causing over half of all foodborne illnesses.
Norovirus is the leading cause of illnesses from contaminated food in the United States, causing over half of all foodborne illnesses. CDC monitors the spread of norovirus strains through CaliciNet, a surveillance network of state and local public health laboratories coordinated by CDC. With AMD methods, CDC is developing a next-generation sequencing (NGS) protocol to amplify norovirus genetic material from patient stool samples. Several CaliciNet labs have implemented these AMD technologies as well, greatly improving public health’s ability to detect outbreaks faster and speed up development of vaccines to prevent norovirus illnesses and outbreaks.

Cyclosporiasis is a foodborne illness. CDC recommends safe food handling techniques for preparing fresh fruits and vegetables.
Cyclospora cayetanensis is a single-celled parasite that causes an intestinal infection called cyclosporiasis. Cyclosporiasis outbreaks often are associated with imported fresh produce. Identifying the particular food item or ingredient that caused an outbreak can be very challenging because fresh produce might be mixed in a salad or served as a garnish or topping. CDC and other agencies are using AMD methods to develop DNA fingerprinting to distinguish among different strains of C. cayetanensis that causes cyclosporiasis. These tools could help link cases of cyclosporiasis to each other and to particular types of produce, which help public health officials investigate and prevent cases and outbreaks of Cyclospora infection.