Use of an Inferred PFGE Algorithm, Emerging Infections Program/Active Bacterial Core (ABCs) Surveillance Invasive MRSA Project
Update December 4, 2009
In 2008, Centers for Disease Control and Prevention’s (CDC’s) Division of Healthcare Quality Promotion developed an algorithm to infer PFGE types among MRSA isolates submitted from the ABCs Invasive MRSA Surveillance Program. This algorithm includes laboratory findings from the past three years of ABC’s MRSA surveillance, in which over 3,500 MRSA isolates (including PFGE) have undergone complete molecular characterization. Very strong associations between specific combinations of phenotypic/molecular characteristics and the definitive PFGE results were identified. Combining these characteristics as part of the algorithm to infer a PFGE result is an efficient way to quickly describe the isolate repository in an accurate manner.
The algorithm is based on isolates’ microbiologic characteristics such as Staphylococcal cassette chromosome (SCCmec) type, presence of Panton Valentine Leukocidin (PVL) and Toxin Shock Syndrome Toxin (TSST) genes, and antimicrobial susceptibility results (Figure 1). All isolates must undergo testing for these characteristics to obtain an inferred PFGE pattern; those with pending testing results are assigned a status of “inferred PFGE pending.” Some isolates have testing results that, when applied to the algorithm, result in no inferred pattern; these isolates must undergo definitive PFGE (usually result in PFGE USA600, USA700, USA800, USA1100, etc.) (Table 1).
A random sample of the isolates that had PFGE inferred will also undergo definitive PFGE testing each year to ensure ongoing accuracy of the inferred PFGE type (Table 2).
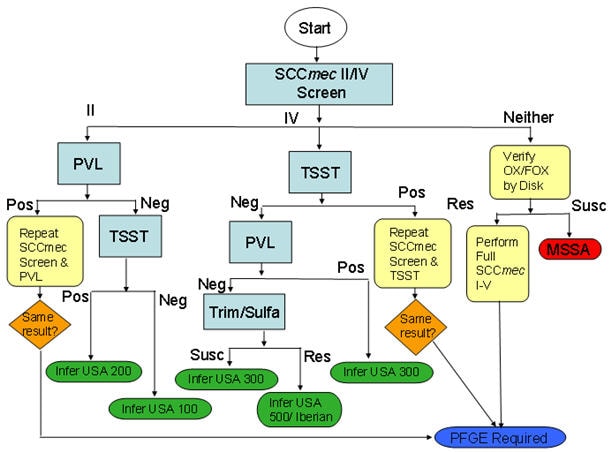
Figure 1: Inferred PFGE Decision Algorithm, ABCs Invasive MRSA Isolates
| Inferred PFGE Type | Epidemiological Classification N (%) | ||||
|---|---|---|---|---|---|
| HO~ (N=247) |
HACO^ (N=595) |
CA† (N=152) |
Unknown‡ (N=11) |
Total (N=1005) |
|
| USA 100 | 177 (71.7) | 363 (61.0) | 46 (30.3) | 5 (45.5) | 591 (58.8) |
| USA 200 | 0 (0) | 1 (0.2) | 0 (0) | 0 (0) | 1 (0.1) |
| USA 300 | 48 (19.4) | 157 (26.4) | 103 (67.8) | 4 (36.4) | 312 (31.0) |
| USA 500/IBERIAN | 22 (8.9) | 64 (10.8) | 2 (1.3) | 0 (0) | 88 (8.8) |
| Direct PFGE Required* | 0 (0) | 10 (16.7) | 1 (0.7) | 2 (18.2) | 13 (1.0) |
~ Hospital Onset Case (HO): a case-patient with infection identified >2 days after hospital admission.
^ Healthcare Associated Community Onset Case (HACO): a case-patient with infection identified ≤2 days after admission to the hospital or had one or more of the following: 1) history of hospitalization, surgery, dialysis or residence of long term care facility in the previous year or 2) the presences of a central vascular catheter.
† Community Associated Case (CA): a case who did not meet any of the above mentioned criteria.
‡ Epidemiological classification could not be determined on the patient after chart review.
*A total of 3 isolates, after completion of the inferred PFGE algorithm, have undergone direct PFGE. 1 isolate was USA 100 and 2 were USA 300. Not represented in this table.
| Inferred PFGE Type | N | Observed PFGE | Details on Incorrect Inference | |
|---|---|---|---|---|
| Correct Inference N (%) |
Incorrect Inference N (%) |
|||
| USA100 | 165 | 163 (98) | 2 (2) | 1=USA700
1=USA800 |
| USA200 | 1 | 0 | 1 (100) | 1=USA100 |
| USA300 | 75 | 65 (86.7) | 10 (13.3) | 6=USA800 1=USA400 1=USA1100 1=IBERIAN |
| USA500/Iberian | 25 | 24 (96.3) | 1 (4) | 1=USA300 |
| None of the above* | 13 | 12 (92.3) | 1 (7.7) | |
| Total | 279 | 264 (94.6%) | 15 (5.4%) | ———- |
Kappa for concurrence of inferred PFGE vs. Observed PFGE = 0.907 (0.86-0.95)
*PFGE is required on these isolates